Crystal Structure Analysis and Conformational Epitope Mutation of Triosephosphate Isomerase, a Mud Crab Allergen.
Xia, F., Li, M.S., Liu, Q.M., Liu, M., Yang, Y., Cao, M.J., Chen, G.X., Jin, T., Liu, G.M.(2019) J Agric Food Chem 67: 12918-12926
- PubMed: 31668066
- DOI: https://doi.org/10.1021/acs.jafc.9b05279
- Primary Citation of Related Structures:
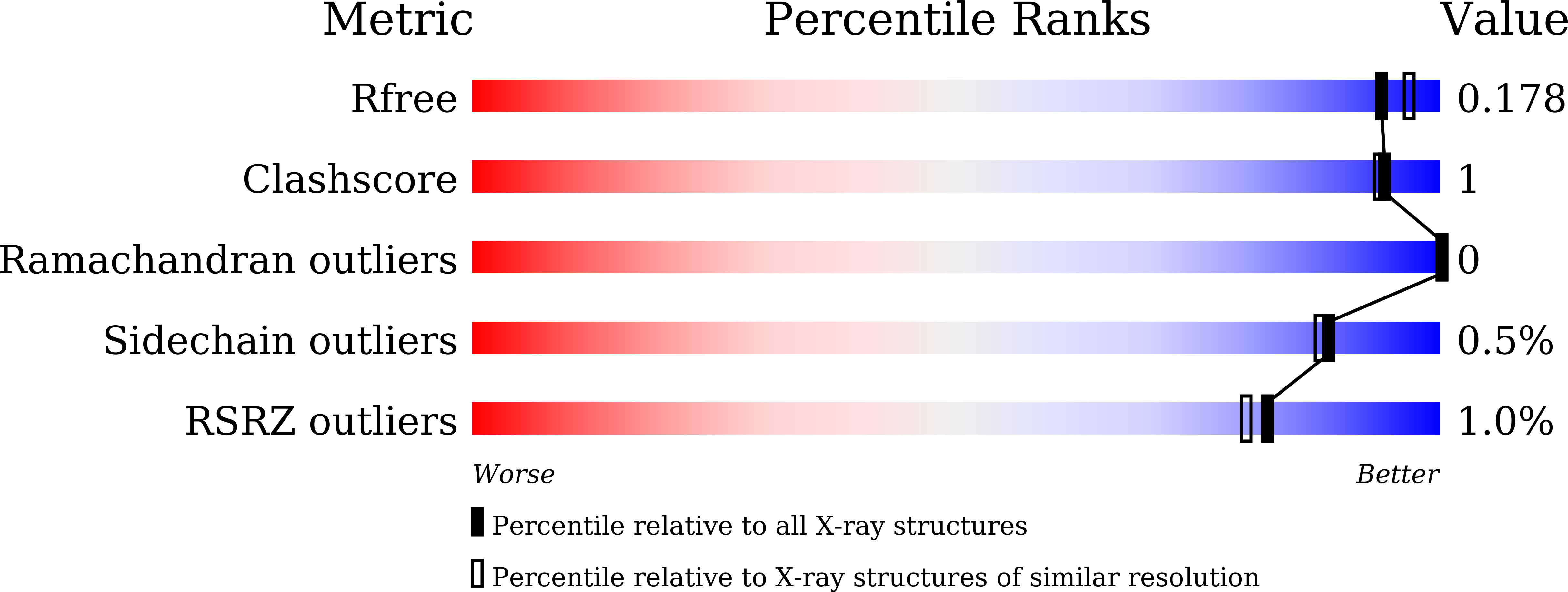
6JOX - PubMed Abstract:
The triosephosphate isomerase (TIM), Scy p 8, is a crab allergen and shows cross-reactivity in the shellfish. Here, recombinant Scy p 8 was expressed, and its crystal structure was determined at a resolution of 1.8 Å. The three-dimensional structure of Scy p 8 is primarily composed of a (β/α) 8 -barrel motif prototype. Additionally, Scy p 8 showed cross-reactivity with high sequential and secondary structural identity among TIMs from shellfish species. The site-directed mutagenesis of critical amino acids of conformational epitopes was carried out, and the mutants of Trp 168 and Lys 237 to Ala reduced immunoglobulin E (IgE)-binding activity by approximately 30%, compared with wild-type TIM in an inhibition ELISA; however, it still induced basophil activation despite the interpatient variability between patients. These results can help to provide an accurate template for the analysis of the IgE binding and establish meaningful relationships between structure and allergenicity.
Organizational Affiliation:
College of Food and Biological Engineering, Xiamen Key Laboratory of Marine Functional Food, Fujian Provincial Engineering Technology Research Center of Marine Functional Food, Fujian Collaborative Innovation Center for Exploitation and Utilization of Marine Biological Resources , Jimei University , Xiamen , Fujian 361021 , China.