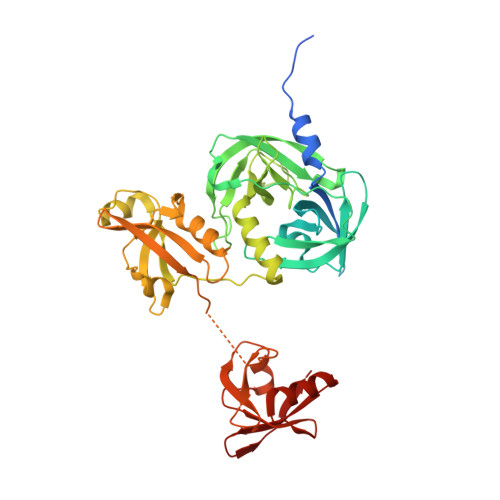
Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Cho, H., Choi, Y., Min, K., Son, J.B., Park, H., Lee, H.H., Kim, S.(2020) Commun Biol 3: 547-547
- PubMed: 33005001
- DOI: https://doi.org/10.1038/s42003-020-01266-9
- Primary Citation of Related Structures:
6JJK, 6JJL, 6JJO - PubMed Abstract:
Rising antibiotic resistance urgently begs for novel targets and strategies for antibiotic discovery. Here, we report that over-activation of the periplasmic DegP protease, a member of the highly conserved HtrA family, can be a viable strategy for antibiotic development. We demonstrate that tripodal peptidyl compounds that mimic DegP-activating lipoprotein variants allosterically activate DegP and inhibit the growth of an Escherichia coli strain with a permeable outer membrane in a DegP-dependent fashion. Interestingly, these compounds inhibit bacterial growth at a temperature at which DegP is not essential for cell viability, mainly by over-proteolysis of newly synthesized proteins. Co-crystal structures show that the peptidyl arms of the compounds bind to the substrate-binding sites of DegP. Overall, our results represent an intriguing example of killing bacteria by activating a non-essential enzyme, and thus expand the scope of antibiotic targets beyond the traditional essential proteins or pathways.
Organizational Affiliation:
Department of Chemistry, Seoul National University, 1 Gwanak-ro, Gwanak-gu, Seoul, 08826, South Korea.