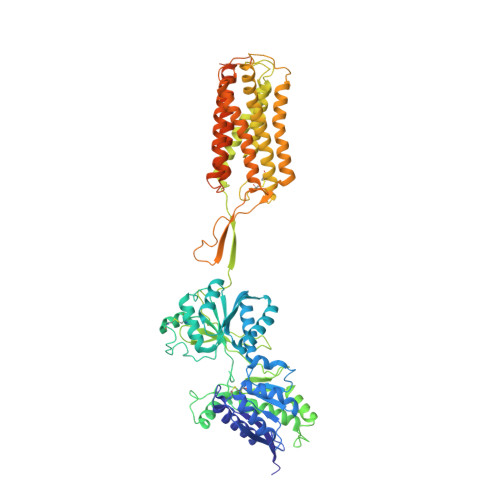
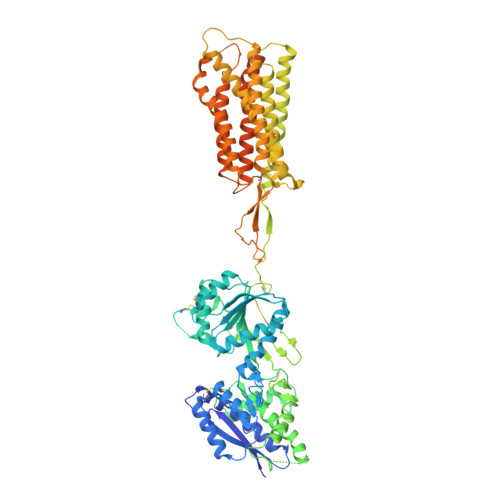
Structural basis of the activation of a metabotropic GABA receptor.
Shaye, H., Ishchenko, A., Lam, J.H., Han, G.W., Xue, L., Rondard, P., Pin, J.P., Katritch, V., Gati, C., Cherezov, V.(2020) Nature 584: 298-303
- PubMed: 32555460
- DOI: https://doi.org/10.1038/s41586-020-2408-4
- Primary Citation of Related Structures:
6UO8, 6UO9, 6UOA, 6VJM - PubMed Abstract:
Metabotropic γ-aminobutyric acid receptors (GABA B ) are involved in the modulation of synaptic responses in the central nervous system and have been implicated in neuropsychological conditions that range from addiction to psychosis 1 . GABA B belongs to class C of the G-protein-coupled receptors, and its functional entity comprises an obligate heterodimer that is composed of the GB1 and GB2 subunits 2 . Each subunit possesses an extracellular Venus flytrap domain, which is connected to a canonical seven-transmembrane domain. Here we present four cryo-electron microscopy structures of the human full-length GB1-GB2 heterodimer: one structure of its inactive apo state, two intermediate agonist-bound forms and an active form in which the heterodimer is bound to an agonist and a positive allosteric modulator. The structures reveal substantial differences, which shed light on the complex motions that underlie the unique activation mechanism of GABA B . Our results show that agonist binding leads to the closure of the Venus flytrap domain of GB1, triggering a series of transitions, first rearranging and bringing the two transmembrane domains into close contact along transmembrane helix 6 and ultimately inducing conformational rearrangements in the GB2 transmembrane domain via a lever-like mechanism to initiate downstream signalling. This active state is stabilized by a positive allosteric modulator binding at the transmembrane dimerization interface.
Organizational Affiliation:
Bridge Institute, USC Michelson Center for Convergent Biosciences, University of Southern California, Los Angeles, CA, USA.