Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Gilan, O., Rioja, I., Knezevic, K., Bell, M.J., Yeung, M.M., Harker, N.R., Lam, E.Y.N., Chung, C.W., Bamborough, P., Petretich, M., Urh, M., Atkinson, S.J., Bassil, A.K., Roberts, E.J., Vassiliadis, D., Burr, M.L., Preston, A.G.S., Wellaway, C., Werner, T., Gray, J.R., Michon, A.M., Gobbetti, T., Kumar, V., Soden, P.E., Haynes, A., Vappiani, J., Tough, D.F., Taylor, S., Dawson, S.J., Bantscheff, M., Lindon, M., Drewes, G., Demont, E.H., Daniels, D.L., Grandi, P., Prinjha, R.K., Dawson, M.A.(2020) Science 368: 387-394
- PubMed: 32193360
- DOI: https://doi.org/10.1126/science.aaz8455
- Primary Citation of Related Structures:
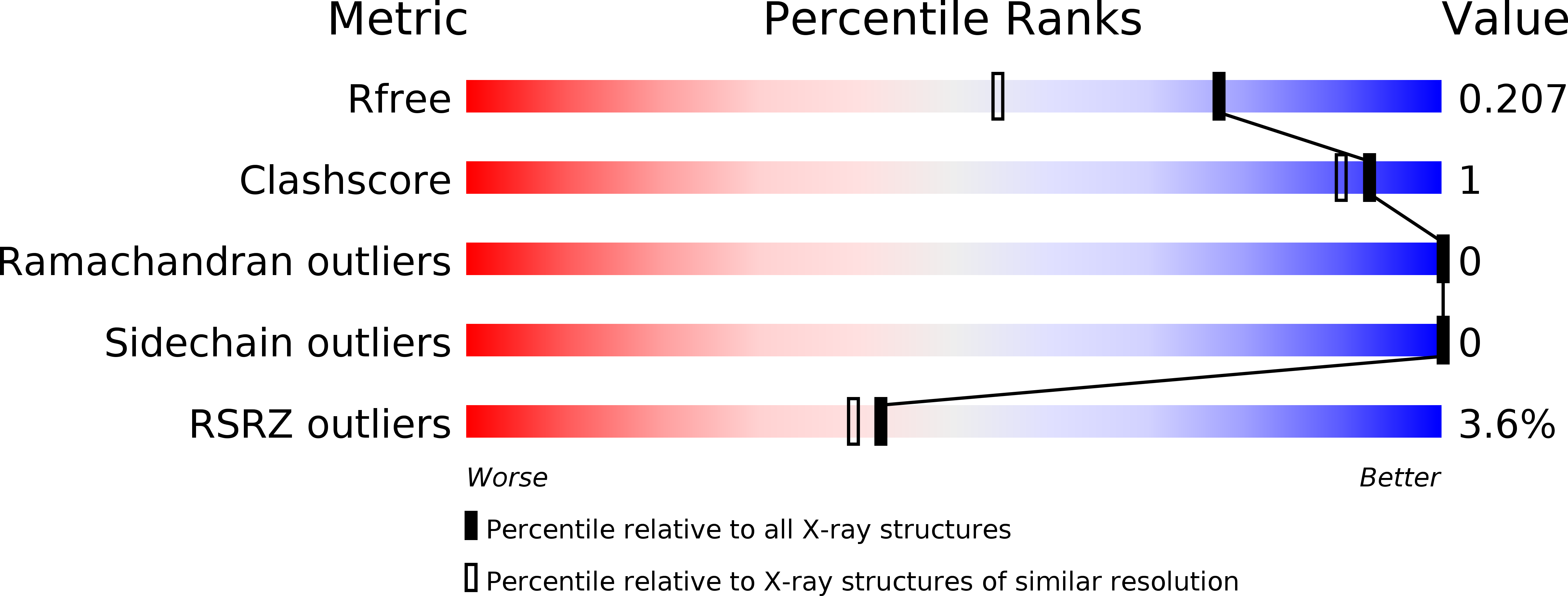
6SWN, 6SWO, 6SWP, 6SWQ - PubMed Abstract:
The two tandem bromodomains of the BET (bromodomain and extraterminal domain) proteins enable chromatin binding to facilitate transcription. Drugs that inhibit both bromodomains equally have shown efficacy in certain malignant and inflammatory conditions. To explore the individual functional contributions of the first (BD1) and second (BD2) bromodomains in biology and therapy, we developed selective BD1 and BD2 inhibitors. We found that steady-state gene expression primarily requires BD1, whereas the rapid increase of gene expression induced by inflammatory stimuli requires both BD1 and BD2 of all BET proteins. BD1 inhibitors phenocopied the effects of pan-BET inhibitors in cancer models, whereas BD2 inhibitors were predominantly effective in models of inflammatory and autoimmune disease. These insights into the differential requirement of BD1 and BD2 for the maintenance and induction of gene expression may guide future BET-targeted therapies.
Organizational Affiliation:
Peter MacCallum Cancer Centre, Melbourne, VIC, Australia.