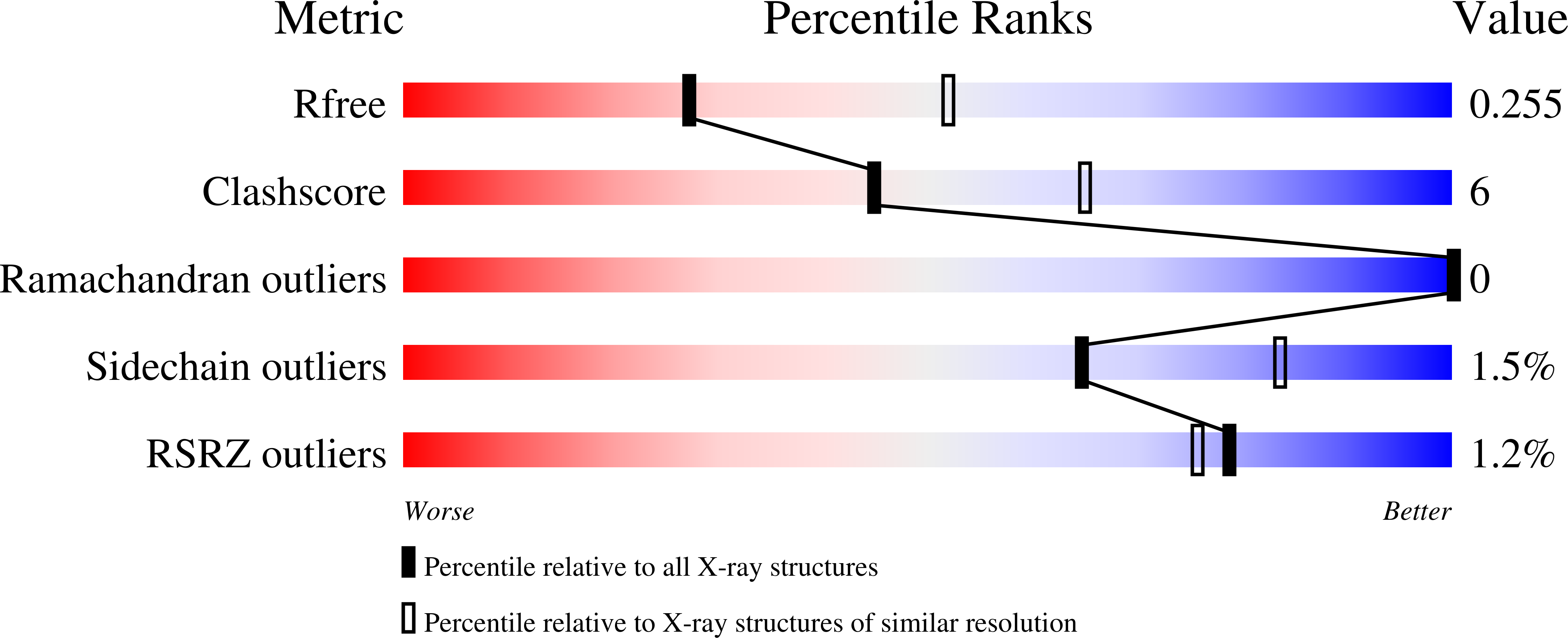
No magnesium is needed for binding of the stimulator of interferon genes to cyclic dinucleotides.
Smola, M., Birkus, G., Boura, E.(2019) Acta Crystallogr F Struct Biol Commun 75: 593-598
- PubMed: 31475926
- DOI: https://doi.org/10.1107/S2053230X19010999
- Primary Citation of Related Structures:
6S86 - PubMed Abstract:
Stimulator of interferon genes (STING) binds cyclic dinucleotides (CDNs), which induce a large conformational change of the protein. The structural basis of activation of STING by CDNs is rather well understood. Unliganded STING forms an open dimer that undergoes a large conformational change (∼10 Å) to a closed conformation upon the binding of a CDN molecule. This event activates downstream effectors of STING and subsequently leads to activation of the type 1 interferon response. However, a previously solved structure of STING with 3',3'-c-di-GMP shows Mg atoms mediating the interaction of STING with this CDN. Here, it is shown that no Mg atoms are needed for this interaction; in fact, magnesium can in some cases obstruct the binding of a CDN to STING.
Organizational Affiliation:
Gilead Sciences Research Centre at IOCB Prague, Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences, Flemingovo nam. 2, 166 10 Prague, Czech Republic.