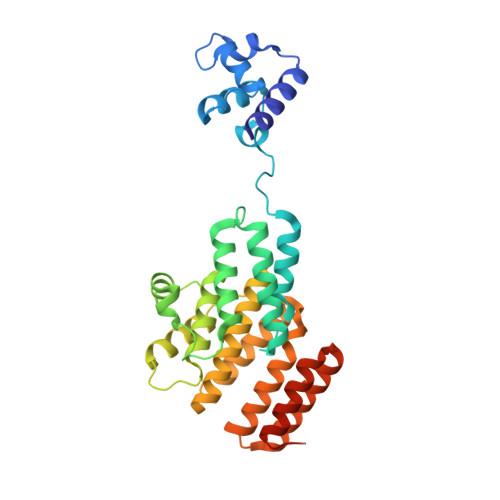
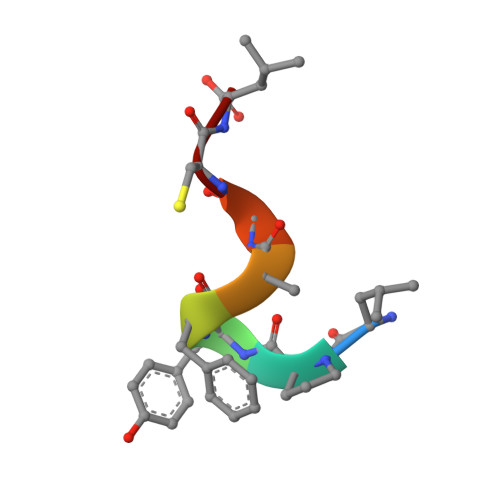
Structural Insights into Streptococcal Competence Regulation by the Cell-to-Cell Communication System ComRS.
Talagas, A., Fontaine, L., Ledesma-Garca, L., Mignolet, J., Li de la Sierra-Gallay, I., Lazar, N., Aumont-Nicaise, M., Federle, M.J., Prehna, G., Hols, P., Nessler, S.(2016) PLoS Pathog 12: e1005980-e1005980
- PubMed: 27907189
- DOI: https://doi.org/10.1371/journal.ppat.1005980
- Primary Citation of Related Structures:
5JUB, 5JUF - PubMed Abstract:
In Gram-positive bacteria, cell-to-cell communication mainly relies on extracellular signaling peptides, which elicit a response either indirectly, by triggering a two-component phosphorelay, or directly, by binding to cytoplasmic effectors. The latter comprise the RNPP family (Rgg and original regulators Rap, NprR, PrgX and PlcR), whose members regulate important bacterial processes such as sporulation, conjugation, and virulence. RNPP proteins are increasingly considered as interesting targets for the development of new antibacterial agents. These proteins are characterized by a TPR-type peptide-binding domain, and except for Rap proteins, also contain an N-terminal HTH-type DNA-binding domain and display a transcriptional activity. Here, we elucidate the structure-function relationship of the transcription factor ComR, a new member of the RNPP family, which positively controls competence for natural DNA transformation in streptococci. ComR is directly activated by the binding of its associated pheromone XIP, the mature form of the comX/sigX-inducing-peptide ComS. The crystal structure analysis of ComR from Streptococcus thermophilus combined with a mutational analysis and in vivo assays allows us to propose an original molecular mechanism of the ComR regulation mode. XIP-binding induces release of the sequestered HTH domain and ComR dimerization to allow DNA binding. Importantly, we bring evidence that this activation mechanism is conserved and specific to ComR orthologues, demonstrating that ComR is not an Rgg protein as initially proposed, but instead constitutes a new member of the RNPP family. In addition, identification of XIP and ComR residues important for competence activation constitutes a crucial step towards the design of antagonistic strategies to control gene exchanges among streptococci.
Organizational Affiliation:
Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Univ. Paris-Sud, Université Paris-Saclay, Gif-sur-Yvette cedex, France.