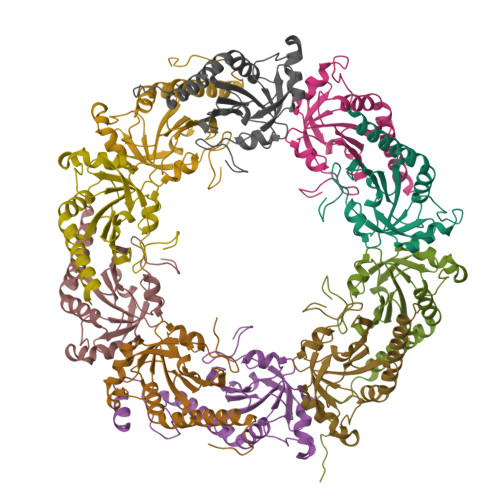
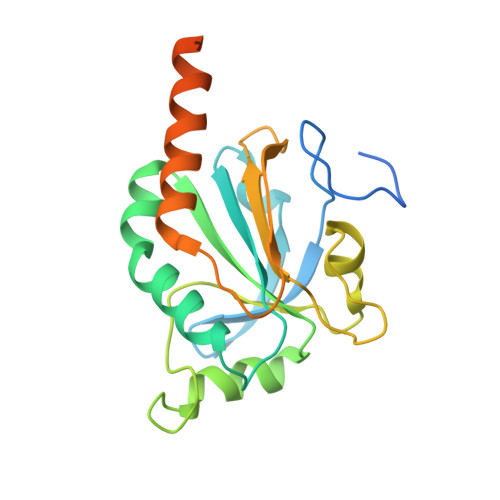
Structure of TSA2 reveals novel features of the active-site loop of peroxiredoxins.
Nielsen, M.H., Kidmose, R.T., Jenner, L.B.(2016) Acta Crystallogr D Struct Biol 72: 158-167
- PubMed: 26894543
- DOI: https://doi.org/10.1107/S2059798315023815
- Primary Citation of Related Structures:
5DVB, 5EPT - PubMed Abstract:
Saccharomyces cerevisiae TSA2 belongs to the family of typical 2-Cys peroxiredoxins, a ubiquitously expressed family of redox-active enzymes that utilize a conserved peroxidatic cysteine to reduce peroxides. Typical 2-Cys peroxiredoxins have been shown to be involved in protection against oxidative stress and in hydrogen peroxide signalling. Furthermore, several 2-Cys peroxiredoxins, including S. cerevisiae TSA1 and TSA2, are able to switch to chaperone activity upon hyperoxidation of their peroxidatic cysteine. This makes the sensitivity to hyperoxidation of the peroxidatic cysteine a very important determinant for the cellular function of a peroxiredoxin under different cellular conditions. Typical 2-Cys peroxiredoxins exist as dimers, and in the course of the reaction the peroxidatic cysteine forms a disulfide with a resolving cysteine located in the C-terminus of its dimeric partner. This requires a local unfolding of the active site and the C-terminus. The balance between the fully folded and locally unfolded conformations is of key importance for the reactivity and sensitivity to hyperoxidation of the different peroxiredoxins. Here, the structure of a C48S mutant of TSA2 from S. cerevisiae that mimics the reduced state of the peroxidatic cysteine has been determined. The structure reveals a novel conformation for the strictly conserved Pro41, which is likely to affect the delicate balance between the fully folded and locally unfolded conformations of the active site, and therefore the reactivity and the sensitivity to hyperoxidation. Furthermore, the structure also explains the observed difference in the pKa values of the peroxidatic cysteines of S. cerevisiae TSA1 and TSA2 despite their very high sequence identity.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Aarhus University, Gustav Wieds Vej 10C, 8000 Aarhus, Denmark.