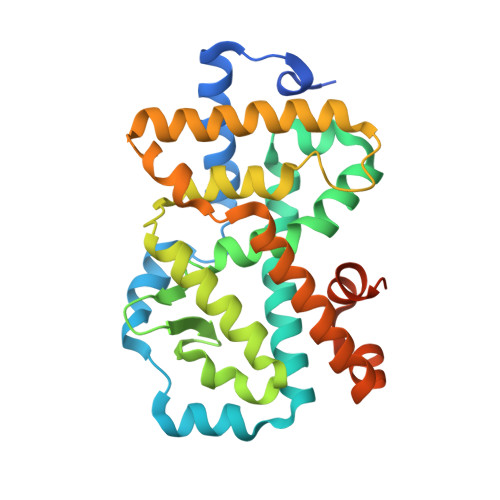
Potent and Orally Bioavailable Inverse Agonists of ROR gamma t Resulting from Structure-Based Design.
Narjes, F., Xue, Y., von Berg, S., Malmberg, J., Llinas, A., Olsson, R.I., Jirholt, J., Grindebacke, H., Leffler, A., Hossain, N., Lepisto, M., Thunberg, L., Leek, H., Aagaard, A., McPheat, J., Hansson, E.L., Back, E., Tangefjord, S., Chen, R., Xiong, Y., Hongbin, G., Hansson, T.G.(2018) J Med Chem 61: 7796-7813
- PubMed: 30095900
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00783
- Primary Citation of Related Structures:
5NI5, 5NI7, 5NI8, 5NIB, 6ESN, 6FGQ - PubMed Abstract:
Retinoic acid receptor related orphan receptor γt (RORγt), has been identified as the master regulator of T H 17-cell function and development, making it an attractive target for the treatment of autoimmune diseases by a small-molecule approach. Herein, we describe our investigations on a series of 4-aryl-thienyl acetamides, which were guided by insights from X-ray cocrystal structures. Efforts in targeting the cofactor-recruitment site from the 4-aryl group on the thiophene led to a series of potent binders with nanomolar activity in a primary human-T H 17-cell assay. The observation of a DMSO molecule binding in a subpocket outside the LBD inspired the introduction of an acetamide into the benzylic position of these compounds. Hereby, a hydrogen-bond interaction of the introduced acetamide oxygen with the backbone amide of Glu379 was established. This greatly enhanced the cellular activity of previously weakly cell-active compounds. The best compounds combined potent inhibition of IL-17 release with favorable PK in rodents, with compound 32 representing a promising starting point for future investigations.
Organizational Affiliation:
Early Product Development , Pharmaceutical Sciences, IMED Biotech Unit, AstraZeneca , Gothenburg , SE-43183 Mölndal , Sweden.