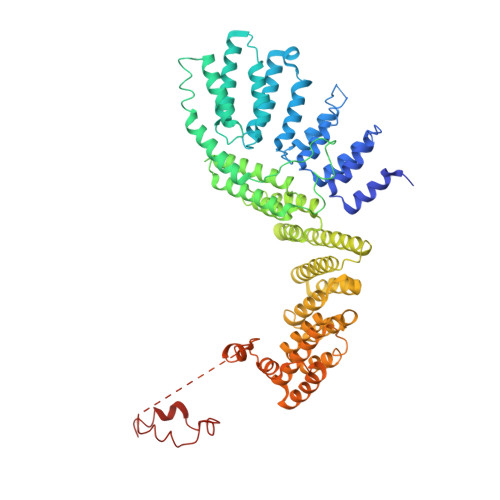
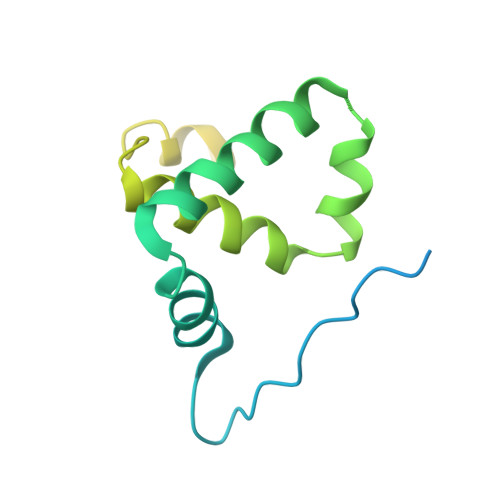
Crystal structure of the Rna14-Rna15 complex.
Paulson, A.R., Tong, L.(2012) RNA 18: 1154-1162
- PubMed: 22513198
- DOI: https://doi.org/10.1261/rna.032524.112
- Primary Citation of Related Structures:
4E6H, 4E85, 4EBA - PubMed Abstract:
A large protein machinery is required for 3'-end processing of mRNA precursors in eukaryotes. Cleavage factor IA (CF IA), a complex in the 3'-end processing machinery in yeast, contains four subunits, Rna14, Rna15, Clp1, and Pcf11. Rna14 has a HAT (half a TPR) domain at the N terminus and a region at the C terminus that mediates interactions with Rna15. Rna15 contains a RNA recognition module (RRM) at the N terminus, followed by a hinge region. These two proteins are homologs of CstF-77 and CstF-64 in the cleavage stimulation factor (CstF) of the mammalian 3'-end processing machinery. We report the first crystal structure of Rna14 in complex with the hinge region of Rna15, and the structures of the HAT domain of Rna14 alone in two different crystal forms. The complex of the C-terminal region of Rna14 with the hinge region of Rna15 does not have strong interactions with the HAT domain of Rna14, and this complex is likely to function independently of the HAT domain. Like CstF-77, the HAT domain of Rna14 is also a tightly associated dimer with a highly elongated shape. However, there are large variations in the organization of this dimer among the Rna14 structures, and there are also significant structural differences to CstF-77. These observations suggest that the HAT domain and especially its dimer may have some inherent conformational variability.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, New York 10027, USA.