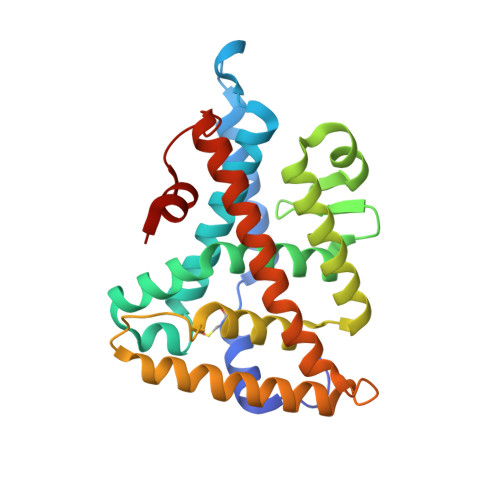
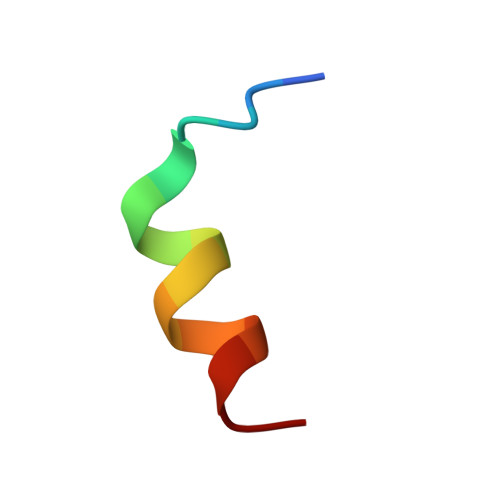
Crystal structure of a Homo sapiens hepatocytic transcription factor hB1F-2 (B1F2) in complex with nuclear receptor subfamily 0 group B member 1 (NR0B1, residues 140-154) from human at 1.86 A resolution
Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell BiologyTo be published.