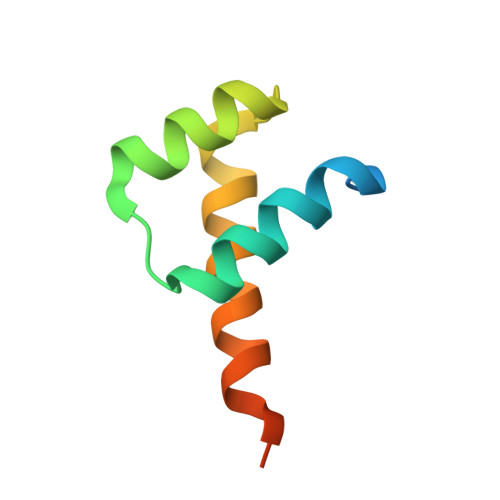
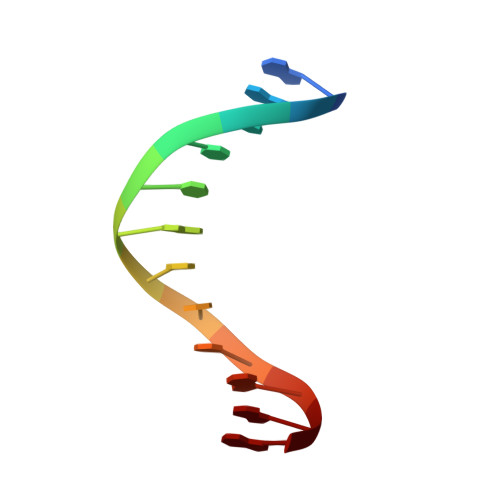
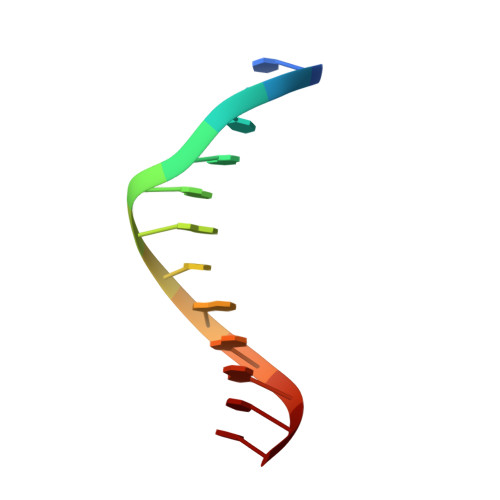
Structure-based discovery of NANOG variant with enhanced properties to promote self-renewal and reprogramming of pluripotent stem cells.
Hayashi, Y., Caboni, L., Das, D., Yumoto, F., Clayton, T., Deller, M.C., Nguyen, P., Farr, C.L., Chiu, H.J., Miller, M.D., Elsliger, M.A., Deacon, A.M., Godzik, A., Lesley, S.A., Tomoda, K., Conklin, B.R., Wilson, I.A., Yamanaka, S., Fletterick, R.J.(2015) Proc Natl Acad Sci U S A 112: 4666-4671
- PubMed: 25825768
- DOI: https://doi.org/10.1073/pnas.1502855112
- Primary Citation of Related Structures:
4RBO - PubMed Abstract:
NANOG (from Irish mythology Tír na nÓg) transcription factor plays a central role in maintaining pluripotency, cooperating with OCT4 (also known as POU5F1 or OCT3/4), SOX2, and other pluripotency factors. Although the physiological roles of the NANOG protein have been extensively explored, biochemical and biophysical properties in relation to its structural analysis are poorly understood. Here we determined the crystal structure of the human NANOG homeodomain (hNANOG HD) bound to an OCT4 promoter DNA, which revealed amino acid residues involved in DNA recognition that are likely to be functionally important. We generated a series of hNANOG HD alanine substitution mutants based on the protein-DNA interaction and evolutionary conservation and determined their biological activities. Some mutant proteins were less stable, resulting in loss or decreased affinity for DNA binding. Overexpression of the orthologous mouse NANOG (mNANOG) mutants failed to maintain self-renewal of mouse embryonic stem cells without leukemia inhibitory factor. These results suggest that these residues are critical for NANOG transcriptional activity. Interestingly, one mutant, hNANOG L122A, conversely enhanced protein stability and DNA-binding affinity. The mNANOG L122A, when overexpressed in mouse embryonic stem cells, maintained their expression of self-renewal markers even when retinoic acid was added to forcibly drive differentiation. When overexpressed in epiblast stem cells or human induced pluripotent stem cells, the L122A mutants enhanced reprogramming into ground-state pluripotency. These findings demonstrate that structural and biophysical information on key transcriptional factors provides insights into the manipulation of stem cell behaviors and a framework for rational protein engineering.
Organizational Affiliation:
Gladstone Institutes of Cardiovascular Disease, San Francisco, CA 94158;