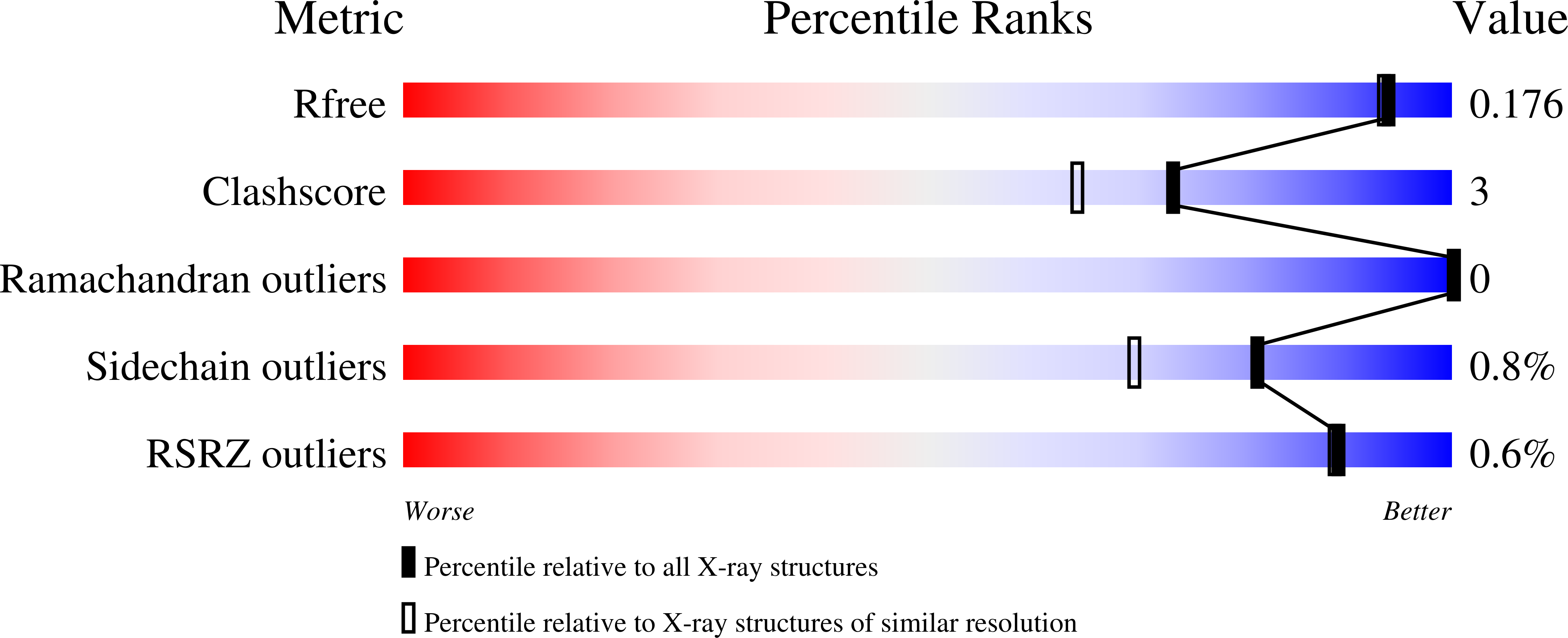
High-resolution crystal structure of the eukaryotic HMP-P synthase (THIC) from Arabidopsis thaliana.
Coquille, S., Roux, C., Mehta, A., Begley, T.P., Fitzpatrick, T.B., Thore, S.(2013) J Struct Biol 184: 438-444
- PubMed: 24161603
- DOI: https://doi.org/10.1016/j.jsb.2013.10.005
- Primary Citation of Related Structures:
4N7Q - PubMed Abstract:
Vitamin B₁ is an essential compound in all organisms acting as a cofactor in key metabolic reactions. It is formed by the condensation of two independently biosynthesized molecules referred to as the pyrimidine and thiazole moieties. In bacteria and plants, the biosynthesis of the pyrimidine moiety, 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate (HMP-P), requires a single enzyme, THIC (HMP-P synthase). The enzyme uses an iron-sulfur cluster as well as a 5'-deoxyadenosyl radical as cofactors to rearrange the 5-amino-imidazole ribonucleotide (AIR) substrate to the pyrimidine ring. So far, the only structure reported is the one from the bacteria Caulobacter crescentus. In an attempt to structurally characterize an eukaryotic HMP-P synthase, we have determined the high-resolution crystal structure of THIC from Arabidopsis thaliana at 1.6 Å. The structure is highly similar to its bacterial counterpart although several loop regions show significant differences with potential implications for the enzymatic properties. Furthermore, we have found a metal ion with octahedral coordination at the same location as a zinc ion in the bacterial enzyme. Our high-resolution atomic model shows a metal ion with multiple coordinated water molecules in the close vicinity of the substrate binding sites and is an important step toward the full characterization of the chemical rearrangement occurring during HMP-P biosynthesis.
Organizational Affiliation:
Department of Molecular Biology, University of Geneva, Geneva 1211, Switzerland. Electronic address: Sandrine.coquille@unige.ch.