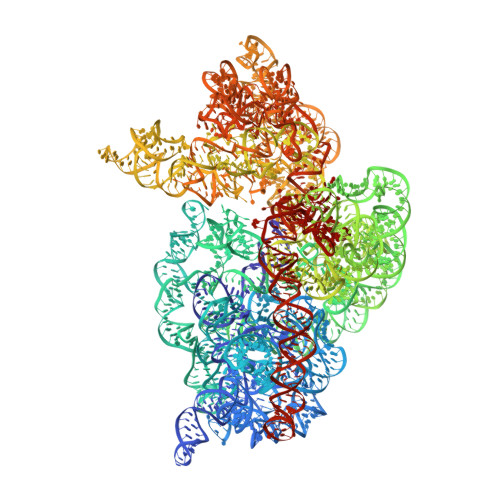
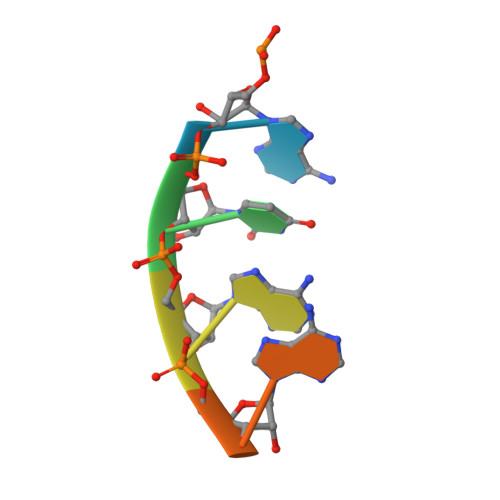
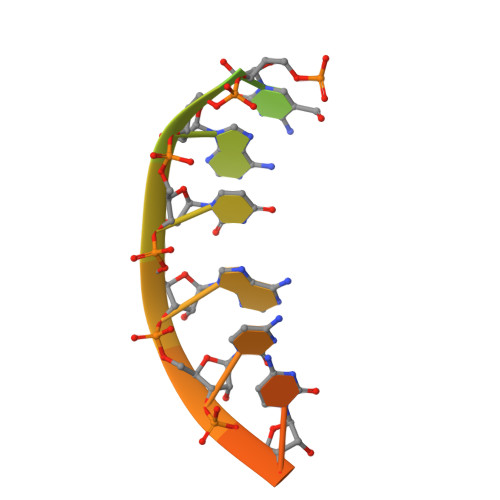
Expanded use of sense codons is regulated by modified cytidines in tRNA.
Cantara, W.A., Murphy, F.V., Demirci, H., Agris, P.F.(2013) Proc Natl Acad Sci U S A 110: 10964-10969
- PubMed: 23781103
- DOI: https://doi.org/10.1073/pnas.1222641110
- Primary Citation of Related Structures:
4GKJ, 4GKK - PubMed Abstract:
Codon use among the three domains of life is not confined to the universal genetic code. With only 22 tRNA genes in mammalian mitochondria, exceptions from the universal code are necessary for proper translation. A particularly interesting deviation is the decoding of the isoleucine AUA codon as methionine by the one mitochondrial-encoded tRNA(Met). This tRNA decodes AUA and AUG in both the A- and P-sites of the metazoan mitochondrial ribosome. Enrichment of posttranscriptional modifications is a commonly appropriated mechanism for modulating decoding rules, enabling some tRNA functions while restraining others. In this case, a modification of cytidine, 5-formylcytidine (f(5)C), at the wobble position-34 of human mitochondrial tRNA(f5CAU)(Met) (hmtRNA(f5CAU)(Met)) enables expanded decoding of AUA, resulting in a deviation in the genetic code. Visualization of the codon•anticodon interaction by X-ray crystallography revealed that recognition of both A and G at the third position of the codon occurs in the canonical Watson-Crick geometry. A modification-dependent shift in the tautomeric equilibrium toward the rare imino-oxo tautomer of cytidine stabilizes the f(5)C34•A base pair geometry with two hydrogen bonds.
Organizational Affiliation:
The RNA Institute, Department of Biological Sciences, University at Albany-State University of New York, Albany, NY 12222, USA.