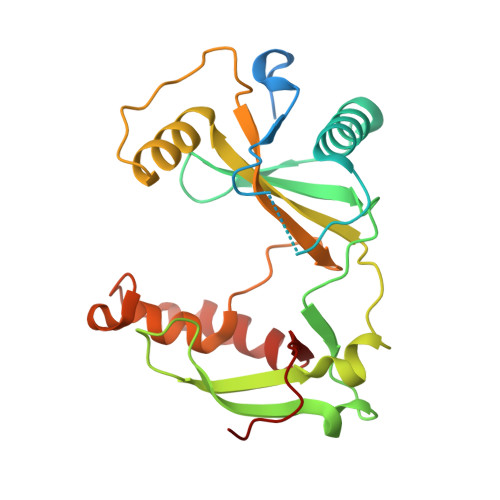
2.35 Angstrom Crystal Structure of Conserved Hypothetical Protein from Toxoplasma gondii ME49.
Minasov, G., Ruan, J., Wawrzak, Z., Shuvalova, L., Ngo, H., Knoll, L., Milligan-Myhre, K., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.