A single mutation at the sheet switch region results in conformational changes favoring lambda6 light-chain fibrillogenesis.
Hernandez-Santoyo, A., del Pozo Yauner, L., Fuentes-Silva, D., Ortiz, E., Rudino-Pinera, E., Sanchez-Lopez, R., Horjales, E., Becerril, B., Rodriguez-Romero, A.(2010) J Mol Biol 396: 280-292
- PubMed: 19941869
- DOI: https://doi.org/10.1016/j.jmb.2009.11.038
- Primary Citation of Related Structures:
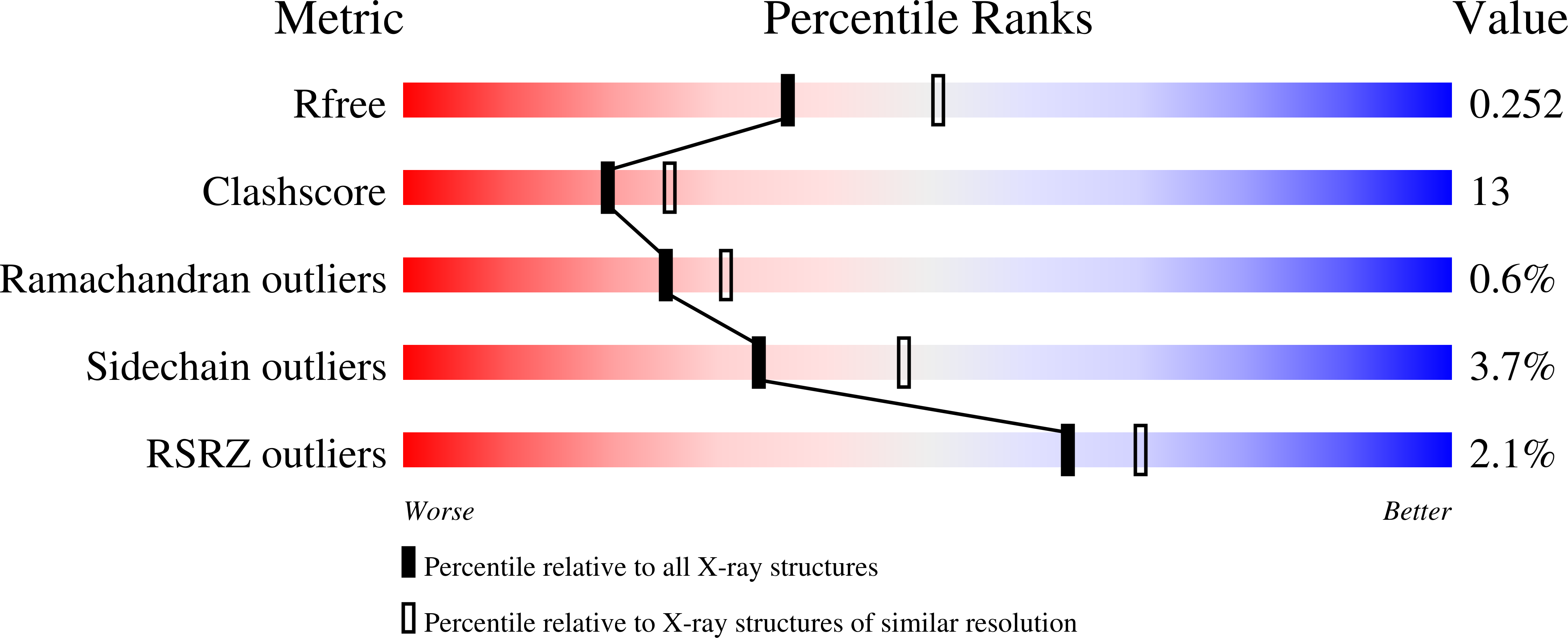
2W0K, 3B5G, 3BDX - PubMed Abstract:
Systemic amyloid light-chain (LC) amyloidosis is a disease process characterized by the pathological deposition of monoclonal LCs in tissue. All LC subtypes are capable of fibril formation although lambda chains, particularly those belonging to the lambda6 type, are overrepresented. Here, we report the thermodynamic and in vitro fibrillogenic properties of several mutants of the lambda6 protein 6aJL2 in which Pro7 and/or His8 was substituted by Ser or Pro. The H8P and H8S mutants were almost as stable as the wild-type protein and were poorly fibrillogenic. In contrast, the P7S mutation decreased the thermodynamic stability of 6aJL2 and greatly enhanced its capacity to form amyloid-like fibrils in vitro. The crystal structure of the P7S mutant showed that the substitution induced both local and long-distance effects, such as the rearrangement of the V(L) (variable region of the light chain)-V(L) interface. This mutant crystallized in two orthorhombic polymorphs, P2(1)2(1)2(1) and C222(1). In the latter, a monomer that was not arranged in the typical Bence-Jones dimer was observed for the first time. Crystal-packing analysis of the C222(1) lattice showed the establishment of intermolecular beta-beta interactions that involved the N-terminus and beta-strand B and that these could be relevant in the mechanism of LC fibril formation. Our results strongly suggest that Pro7 is a key residue in the conformation of the N-terminal sheet switch motif and, through long-distance interactions, is also critically involved in the contacts that stabilized the V(L) interface in lambda6 LCs.
Organizational Affiliation:
Instituto de Química, Universidad Nacional Autónoma de México, Circuito Exterior, Ciudad Universitaria, Mexico DF, Mexico. hersan@servidor.unam.mx