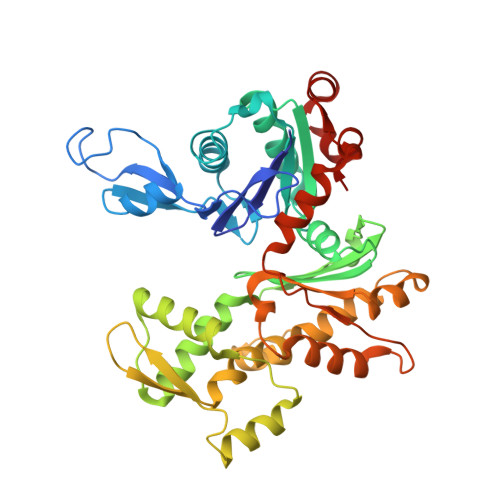
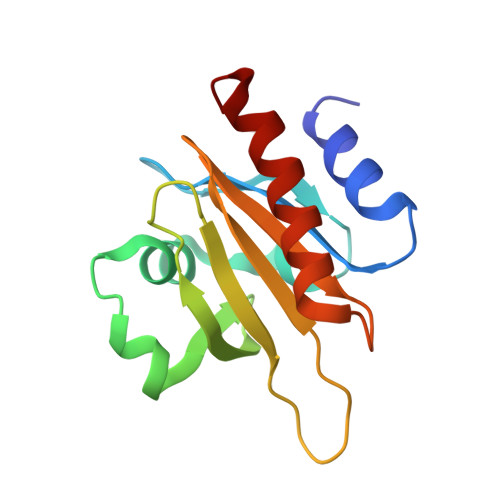
Structural basis for profilin-mediated actin nucleotide exchange.
Porta, J.C., Borgstahl, G.E.(2012) J Mol Biol 418: 103-116
- PubMed: 22366544
- DOI: https://doi.org/10.1016/j.jmb.2012.02.012
- Primary Citation of Related Structures:
3U4L, 3UB5 - PubMed Abstract:
Actin is a ubiquitous eukaryotic protein that is responsible for cellular scaffolding, motility, and division. The ability of actin to form a helical filament is the driving force behind these cellular activities. Formation of a filament depends on the successful exchange of actin's ADP for ATP. Mammalian profilin is a small actin binding protein that catalyzes the exchange of nucleotide and facilitates the addition of an actin monomer to a growing filament. Here, crystal structures of profilin-actin have been determined to show an actively exchanging ATP. Structural analysis shows how the binding of profilin to the barbed end of actin causes a rotation of the small domain relative to the large domain. This conformational change is propagated to the ATP site and causes a shift in nucleotide loops, which in turn causes a repositioning of Ca(2+) to its canonical position as the cleft closes around ATP. Reversal of the solvent exposure of Trp356 is also involved in cleft closure. In addition, secondary calcium binding sites were identified.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, 987696 Nebraska Medical Center, Omaha, NE 68198-7696, USA.