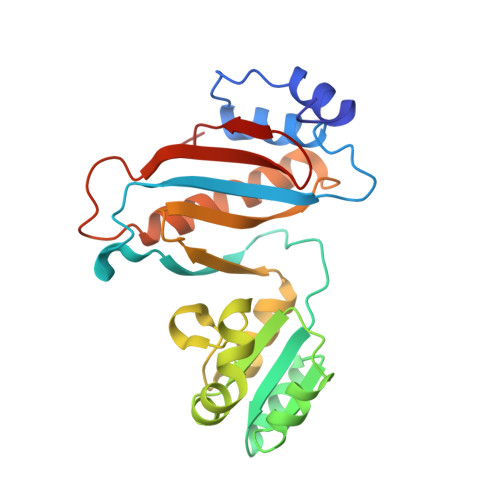
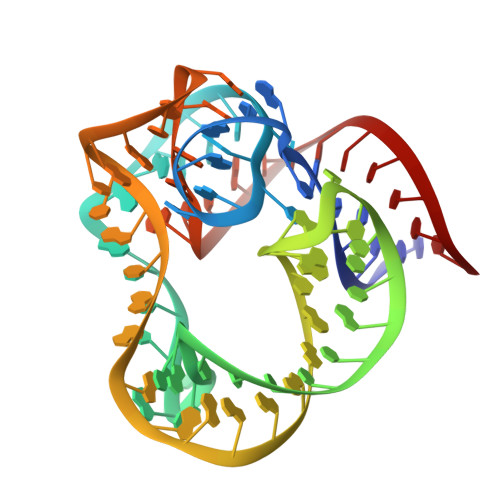
High-resolution crystal structure of the isolated ribosomal L1 stalk.
Tishchenko, S., Gabdulkhakov, A., Nevskaya, N., Sarskikh, A., Kostareva, O., Nikonova, E., Sycheva, A., Moshkovskii, S., Garber, M., Nikonov, S.(2012) Acta Crystallogr D Biol Crystallogr 68: 1051-1057
- PubMed: 22868771
- DOI: https://doi.org/10.1107/S0907444912020136
- Primary Citation of Related Structures:
3U4M - PubMed Abstract:
The crystal structure of the isolated full-length ribosomal L1 stalk, consisting of Thermus thermophilus ribosomal protein L1 in complex with a specific 80-nucleotide fragment of 23S rRNA, has been solved for the first time at high resolution. The structure revealed details of protein-RNA interactions in the L1 stalk. Analysis of the crystal packing enabled the identification of sticky sites on the protein and the 23S rRNA which may be important for ribosome assembly and function. The structure was used to model different conformational states of the ribosome. This approach provides an insight into the roles of domain II of L1 and helix 78 of rRNA in ribosome function.
Organizational Affiliation:
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russian Federation.