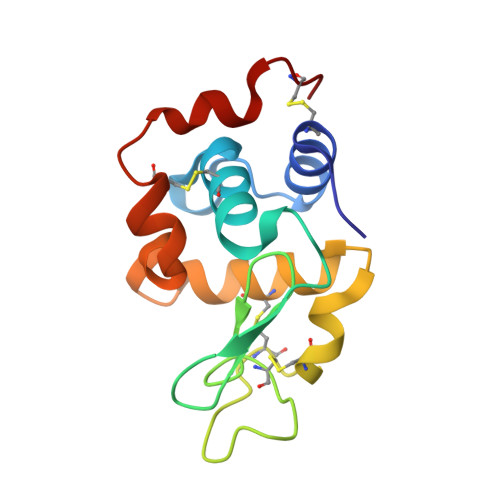
Silver metallation of hen egg white lysozyme: X-ray crystal structure and NMR studies.
Panzner, M.J., Bilinovich, S.M., Youngs, W.J., Leeper, T.C.(2011) Chem Commun (Camb) 47: 12479-12481
- PubMed: 22042312
- DOI: https://doi.org/10.1039/c1cc15908a
- Primary Citation of Related Structures:
3RU5 - PubMed Abstract:
The X-ray crystal structure, NMR binding studies, and enzyme activity of silver(I) metallated hen egg white lysozyme are presented. Primary bonding of silver is observed through His15 with secondary bonding interactions coming from nearby Arg14 and Asp87. A covalently bound nitrate completes a four coordinate binding pocket.
Organizational Affiliation:
Center for Silver Therapeutics Research, Department of Chemistry, The University of Akron, Akron, OH 44325-3601, USA.