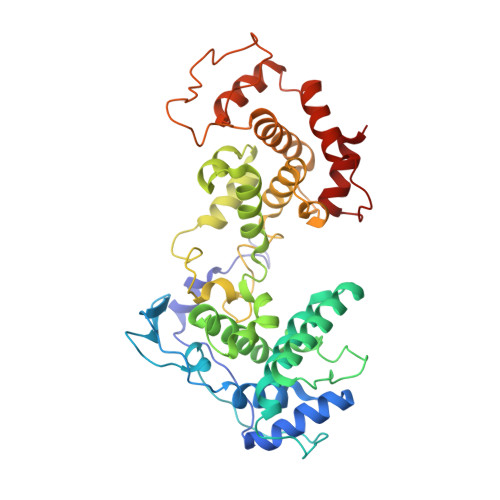
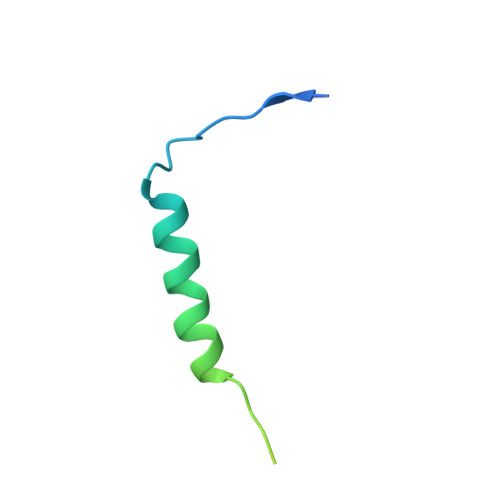
Structure of the Vesicular Stomatitis Virus N0-P Complex
Leyrat, C., Yabukarski, F., Tarbouriech, N., Ribeiro, E.A., Jensen, M.R., Blackledge, M., Ruigrok, R.W., Jamin, M.(2011) PLoS Pathog 7: e1002248-e1002248
- PubMed: 21960769
- DOI: https://doi.org/10.1371/journal.ppat.1002248
- Primary Citation of Related Structures:
3PMK - PubMed Abstract:
Replication of non-segmented negative-strand RNA viruses requires the continuous supply of the nucleoprotein (N) in the form of a complex with the phosphoprotein (P). Here, we present the structural characterization of a soluble, heterodimeric complex between a variant of vesicular stomatitis virus N lacking its 21 N-terminal residues (N(Δ21)) and a peptide of 60 amino acids (P(60)) encompassing the molecular recognition element (MoRE) of P that binds RNA-free N (N(0)). The complex crystallized in a decameric circular form, which was solved at 3.0 Å resolution, reveals how the MoRE folds upon binding to N and competes with RNA binding and N polymerization. Small-angle X-ray scattering experiment and NMR spectroscopy on the soluble complex confirms the binding of the MoRE and indicates that its flanking regions remain flexible in the complex. The structure of this complex also suggests a mechanism for the initiation of viral RNA synthesis.
Organizational Affiliation:
UMI 3265 UJF-EMBL-CNRS Unit of Virus Host Cell Interactions, Grenoble, France.