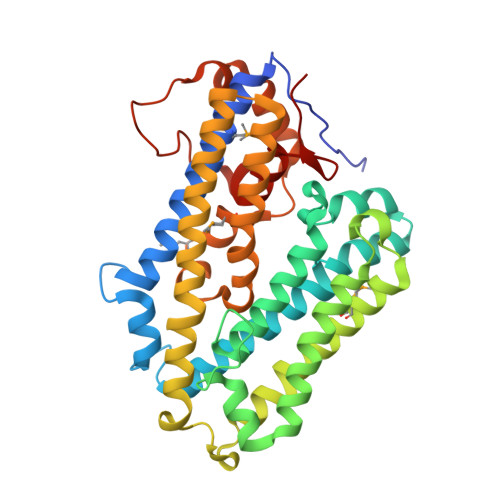
Structural and sequence analysis of imelysin-like proteins implicated in bacterial iron uptake.
Xu, Q., Rawlings, N.D., Farr, C.L., Chiu, H.J., Grant, J.C., Jaroszewski, L., Klock, H.E., Knuth, M.W., Miller, M.D., Weekes, D., Elsliger, M.A., Deacon, A.M., Godzik, A., Lesley, S.A., Wilson, I.A.(2011) PLoS One 6: e21875-e21875
- PubMed: 21799754
- DOI: https://doi.org/10.1371/journal.pone.0021875
- Primary Citation of Related Structures:
3N8U, 3OYV - PubMed Abstract:
Imelysin-like proteins define a superfamily of bacterial proteins that are likely involved in iron uptake. Members of this superfamily were previously thought to be peptidases and were included in the MEROPS family M75. We determined the first crystal structures of two remotely related, imelysin-like proteins. The Psychrobacter arcticus structure was determined at 2.15 Å resolution and contains the canonical imelysin fold, while higher resolution structures from the gut bacteria Bacteroides ovatus, in two crystal forms (at 1.25 Å and 1.44 Å resolution), have a circularly permuted topology. Both structures are highly similar to each other despite low sequence similarity and circular permutation. The all-helical structure can be divided into two similar four-helix bundle domains. The overall structure and the GxHxxE motif region differ from known HxxE metallopeptidases, suggesting that imelysin-like proteins are not peptidases. A putative functional site is located at the domain interface. We have now organized the known homologous proteins into a superfamily, which can be separated into four families. These families share a similar functional site, but each has family-specific structural and sequence features. These results indicate that imelysin-like proteins have evolved from a common ancestor, and likely have a conserved function.
Organizational Affiliation:
Joint Center for Structural Genomics, La Jolla, California, United States of America.