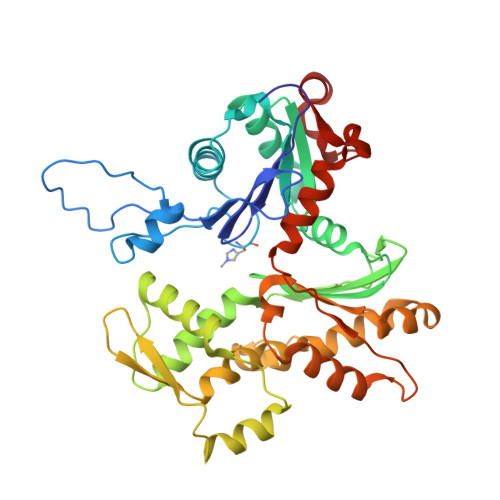
Direct visualization of secondary structures of F-actin by electron cryomicroscopy
Fujii, T., Iwane, A.H., Yanagida, T., Namba, K.(2010) Nature 467: 724-728
- PubMed: 20844487
- DOI: https://doi.org/10.1038/nature09372
- Primary Citation of Related Structures:
3MFP - PubMed Abstract:
F-actin is a helical assembly of actin, which is a component of muscle fibres essential for contraction and has a crucial role in numerous cellular processes, such as the formation of lamellipodia and filopodia, as the most abundant component and regulator of cytoskeletons by dynamic assembly and disassembly (from G-actin to F-actin and vice versa). Actin is a ubiquitous protein and is involved in important biological functions, but the definitive high-resolution structure of F-actin remains unknown. Although a recent atomic model well reproduced X-ray fibre diffraction intensity data from a highly oriented liquid-crystalline sol specimen, its refinement without experimental phase information has certain limitations. Direct visualization of the structure by electron cryomicroscopy, however, has been difficult because it is relatively thin and flexible. Here we report the F-actin structure at 6.6 Å resolution, made obtainable by recent advances in electron cryomicroscopy. The density map clearly resolves all the secondary structures of G-actin, such as α-helices, β-structures and loops, and makes unambiguous modelling and refinement possible. Complex domain motions that open the nucleotide-binding pocket on F-actin formation, specific D-loop and terminal conformations, and relatively tight axial but markedly loose interprotofilament interactions hydrophilic in nature are revealed in the F-actin model, and all seem to be important for dynamic functions of actin.
Organizational Affiliation:
Graduate School of Frontier Biosciences, Osaka University, 1-3 Yamadaoka, Suita, Osaka 565-0871, Japan.