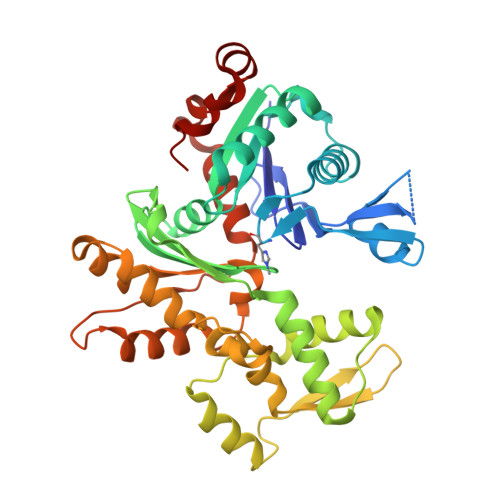
The structure of native G-actin
Wang, H., Robinson, R.C., Burtnick, L.D.(2010) Cytoskeleton (Hoboken) 67: 456-465
- PubMed: 20540085
- DOI: https://doi.org/10.1002/cm.20458
- Primary Citation of Related Structures:
3HBT - PubMed Abstract:
Heat shock proteins act as cytoplasmic chaperones to ensure correct protein folding and prevent protein aggregation. The presence of stoichiometric amounts of one such heat shock protein, Hsp27, in supersaturated solutions of unmodified G-actin leads to crystallization, in preference to polymerization, of the actin. Hsp27 is not evident in the resulting crystal structure. Thus, for the first time, we present the structure of G-actin in a form that is devoid of polymerization-deterring chemical modifications or binding partners, either of which may alter its conformation. The structure contains a calcium ion and ATP within a closed nucleotide-binding cleft, and the D-loop is disordered. This native G-actin structure invites comparison with the current F-actin model in order to understand the structural implications for actin polymerization. In particular, this analysis suggests a mechanism by which the bound cation coordinates conformational change and ATP-hydrolysis.
Organizational Affiliation:
Department of Chemistry and Centre for Blood Research, Life Sciences Institute, University of British Columbia, Vancouver, British Columbia, Canada.