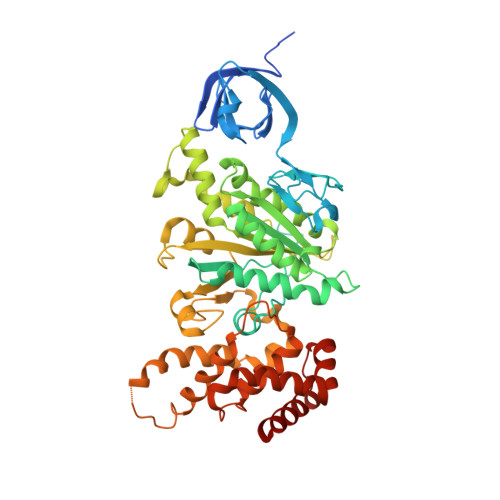
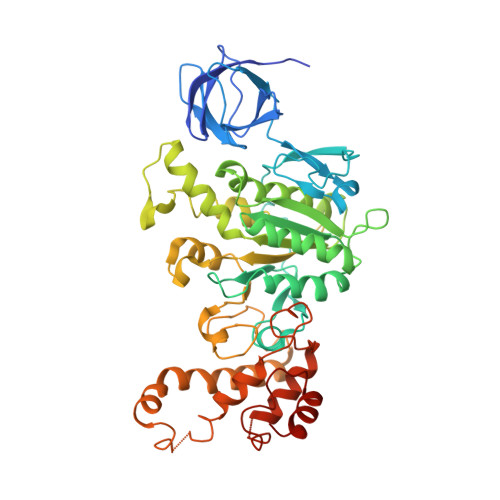
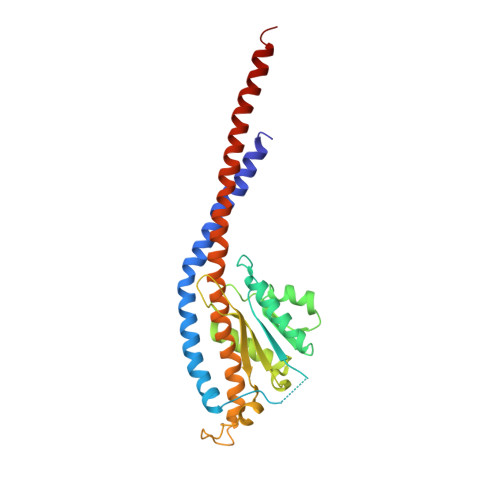
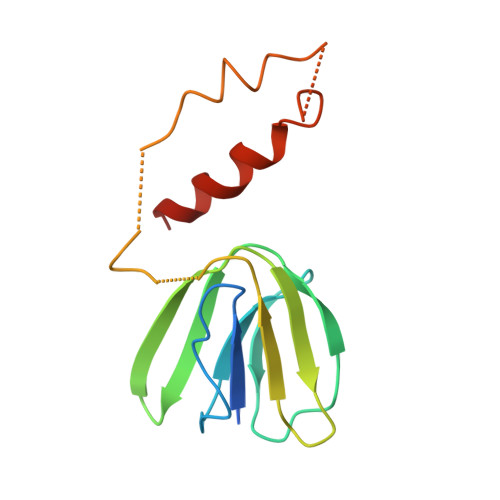
Asymmetric structure of the yeast f1 ATPase in the absence of bound nucleotides.
Kabaleeswaran, V., Shen, H., Symersky, J., Walker, J.E., Leslie, A.G., Mueller, D.M.(2009) J Biol Chem 284: 10546-10551
- PubMed: 19233840
- DOI: https://doi.org/10.1074/jbc.M900544200
- Primary Citation of Related Structures:
3FKS - PubMed Abstract:
The crystal structure of nucleotide-free yeast F(1) ATPase has been determined at a resolution of 3.6 A. The overall structure is very similar to that of the ground state enzyme. In particular, the beta(DP) and beta(TP) subunits both adopt the closed conformation found in the ground state structure despite the absence of bound nucleotides. This implies that interactions between the gamma and beta subunits are as important as nucleotide occupancy in determining the conformational state of the beta subunits. Furthermore, this result suggests that for the mitochondrial enzyme, there is no state of nucleotide occupancy that would result in more than one of the beta subunits adopting the open conformation. The adenine-binding pocket of the beta(TP) subunit is disrupted in the apoenzyme, suggesting that the beta(DP) subunit is responsible for unisite catalytic activity.
Organizational Affiliation:
Rosalind Franklin University of Medicine and Science, The Chicago Medical School, North Chicago, Illinois 60064, USA.