Probing Wavelength Regulation with an Engineered Rhodopsin Mimic and a C15-Retinal Analogue
Lee, K.S., Berbasova, T., Vasileiou, C., Jia, X., Wang, W., Choi, Y., Nossoni, F., Geiger, J.H., Borhan, B.(2012) Chempluschem 77: 273-276
Experimental Data Snapshot
(2012) Chempluschem 77: 273-276
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cellular retinoic acid-binding protein 2 | 137 | Homo sapiens | Mutation(s): 4 Gene Names: CRABP2 | 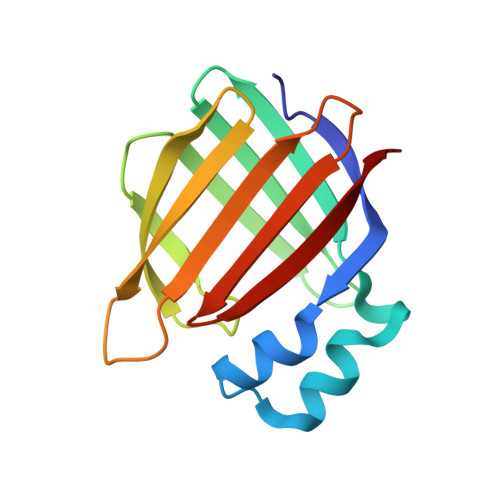 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P29373 (Homo sapiens) Explore P29373 Go to UniProtKB: P29373 | |||||
PHAROS: P29373 GTEx: ENSG00000143320 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P29373 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| B3P Query on B3P | B [auth A] | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C11 H26 N2 O6 HHKZCCWKTZRCCL-UHFFFAOYSA-N |  | ||
| LSR Query on LSR | C [auth A] | 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene C15 H24 KUEVAPFABUUVHS-AYCKBHPDSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 58.468 | α = 90 |
| b = 58.468 | β = 90 |
| c = 104.031 | γ = 120 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |