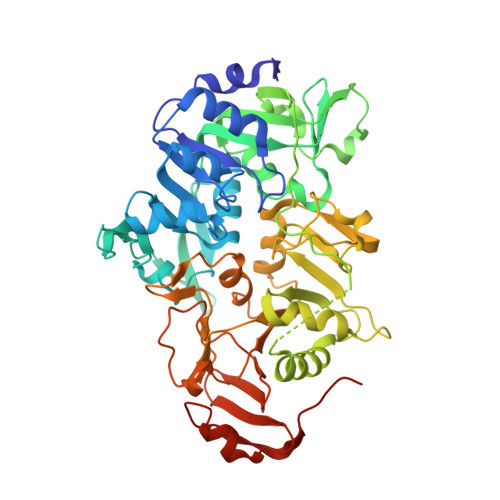
Crystal structure of human 3-oxoacid CoA transferase 1.
Kavanagh, K.L., Shafqat, N., Yue, W.W., Picaud, S., Murray, J.W., Maclean, E.M., von Delft, F., Roos, A.K., Arrowsmith, C.H., Wikstrom, M., Edwards, A.M., Bountra, C., Oppermann, U.To be published.