Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
Han, Q., Cai, T., Tagle, D.A., Robinson, H., Li, J.(2008) Biosci Rep 28: 205-215
- PubMed: 18620547
- DOI: https://doi.org/10.1042/BSR20080085
- Primary Citation of Related Structures:
3DC1 - PubMed Abstract:
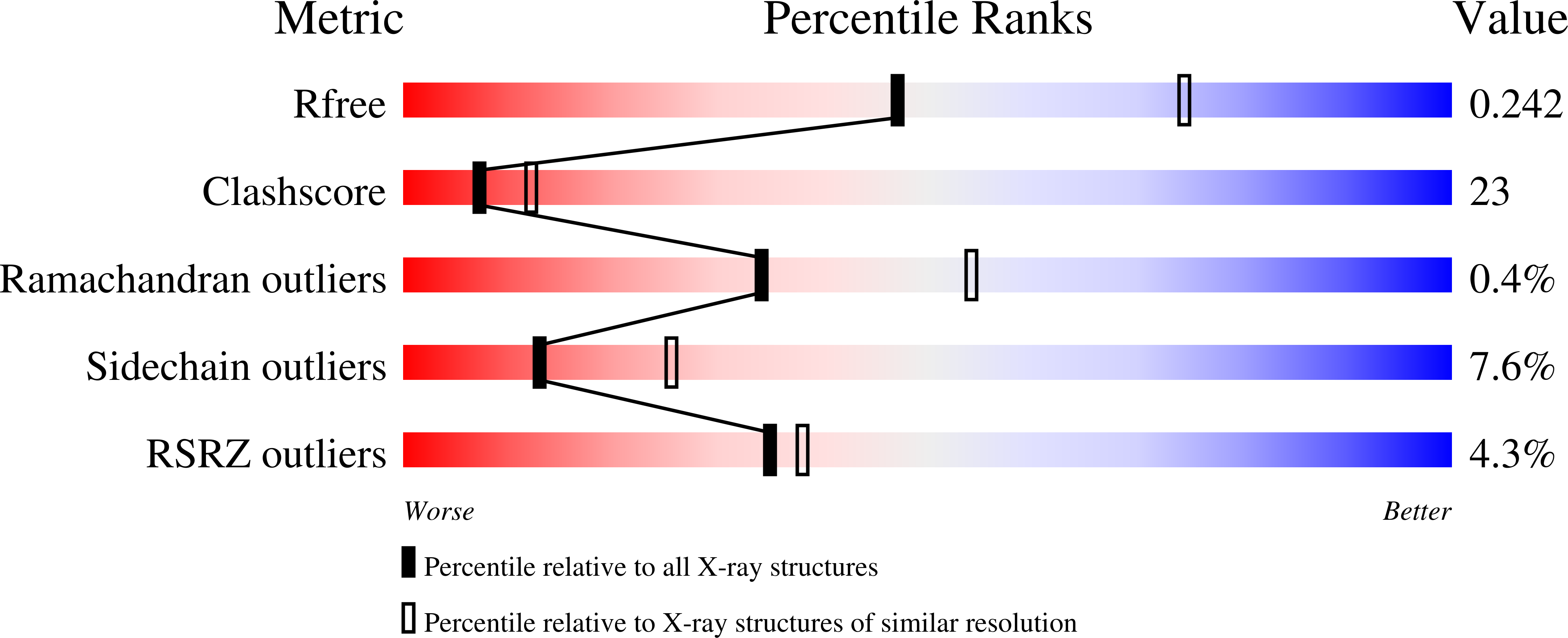
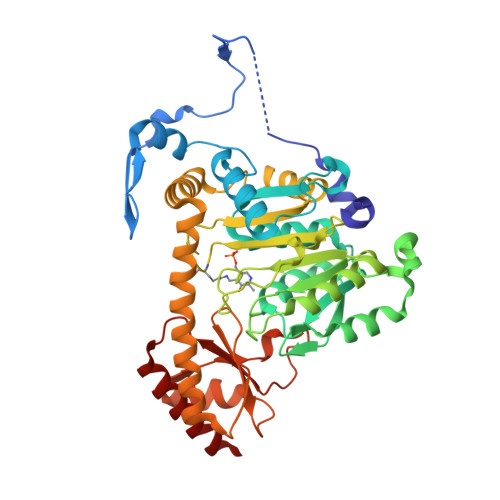
KAT (kynurenine aminotransferase) II is a primary enzyme in the brain for catalysing the transamination of kynurenine to KYNA (kynurenic acid). KYNA is the only known endogenous antagonist of the N-methyl-D-aspartate receptor. The enzyme also catalyses the transamination of aminoadipate to alpha-oxoadipate; therefore it was initially named AADAT (aminoadipate aminotransferase). As an endotoxin, aminoadipate influences various elements of glutamatergic neurotransmission and kills primary astrocytes in the brain. A number of studies dealing with the biochemical and functional characteristics of this enzyme exist in the literature, but a systematic assessment of KAT II addressing its substrate profile and kinetic properties has not been performed. The present study examines the biochemical and structural characterization of a human KAT II/AADAT. Substrate screening of human KAT II revealed that the enzyme has a very broad substrate specificity, is capable of catalysing the transamination of 16 out of 24 tested amino acids and could utilize all 16 tested alpha-oxo acids as amino-group acceptors. Kinetic analysis of human KAT II demonstrated its catalytic efficiency for individual amino-group donors and acceptors, providing information as to its preferred substrate affinity. Structural analysis of the human KAT II complex with alpha-oxoglutaric acid revealed a conformational change of an N-terminal fraction, residues 15-33, that is able to adapt to different substrate sizes, which provides a structural basis for its broad substrate specificity.
Organizational Affiliation:
Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, U.S.A. qianhan@vt.edu