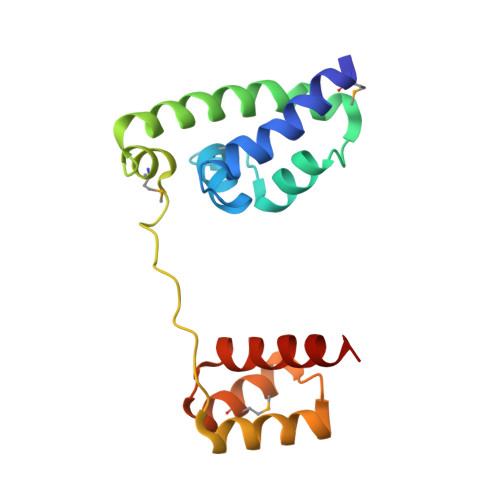
Structure of human Brn-5 transcription factor in complex with CRH gene promoter.
Pereira, J.H., Kim, S.H.(2009) J Struct Biol 167: 159-165
- PubMed: 19450691
- DOI: https://doi.org/10.1016/j.jsb.2009.05.003
- Primary Citation of Related Structures:
3D1N - PubMed Abstract:
The Brn-5 protein, highly expressed in human brain, belongs to the POU family; a class of transcription factors involved in a wide variety of biological processes ranging from programming of embryonic stem cells to cellular housekeeping. This functional diversity is conferred by two DNA-binding subdomains that can assume several configurations due to a bipartite arrangement of POU-specific (POU(S)) and POU-homeo (POU(H)) subdomains separated by a linker region. The crystal structure of human Brn-5 transcription factor in complex with corticotrophin-releasing hormone (CRH) gene promoter reveals an unexpected recognition mode of the protein to its cognate DNA. Moreover, the structure also shows the role of the linker in allowing diverse configurations that can be assumed by the two subdomains.
Organizational Affiliation:
Department of Chemistry, University of California, Berkeley, CA 94720, USA.