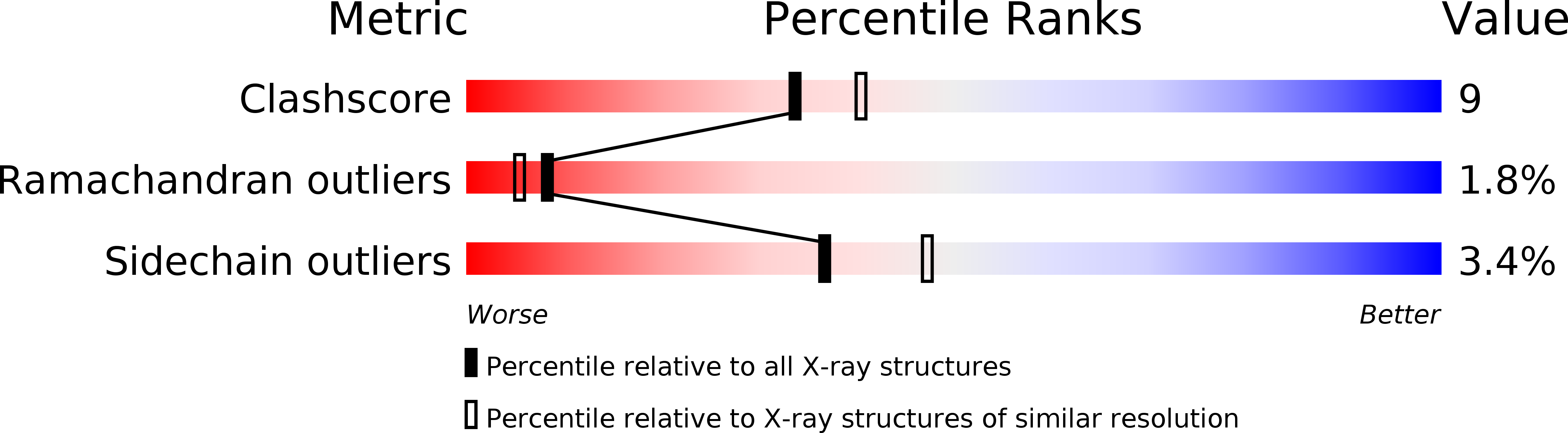Structural Basis for Dimerization of Lap2Alpha, a Component of the Nuclear Lamina.
Bradley, C.M., Jones, S., Huang, Y., Suzuki, Y., Kvaratskhelia, M., Hickman, A.B., Craigie, R., Dyda, F.(2007) Structure 15: 643
- PubMed: 17562312
- DOI: https://doi.org/10.1016/j.str.2007.04.007
- Primary Citation of Related Structures:
2V0X - PubMed Abstract:
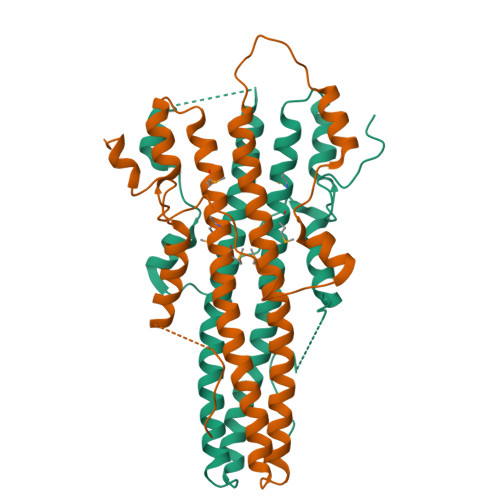
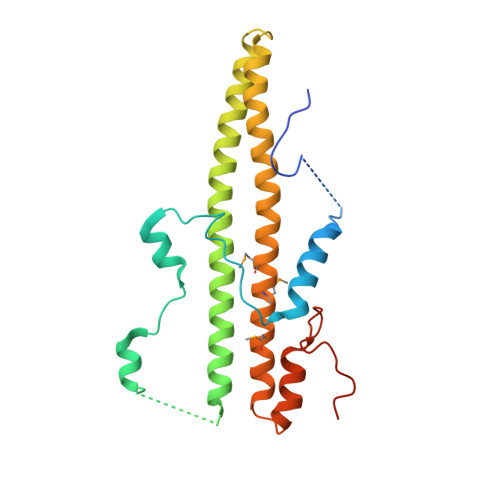
Lamina-associated polypeptides (LAPs) are important components of the nuclear lamina, the dense network of filaments that supports the nuclear envelope and also extends into the nucleoplasm. The main protein constituents of the nuclear lamina are the constitutively expressed B-type lamins and the developmentally regulated A- and C-type lamins. LAP2alpha is the only non-membrane-associated member of the LAP family. It preferentially binds lamin A/C, has been implicated in cell-cycle regulation and chromatin organization, and has also been found to be a component of retroviral preintegration complexes. As an approach to understanding the role of LAP2alpha in cellular pathways, we have determined the crystal structure of the C-terminal domain of LAP2alpha, residues 459-693. The C-terminal domain is dimeric and possesses an extensive four-stranded, antiparallel coiled coil. The surface involved in binding lamin A/C is proposed based on results from alanine-scanning mutagenesis and a solid-phase overlay binding assay.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892, USA.