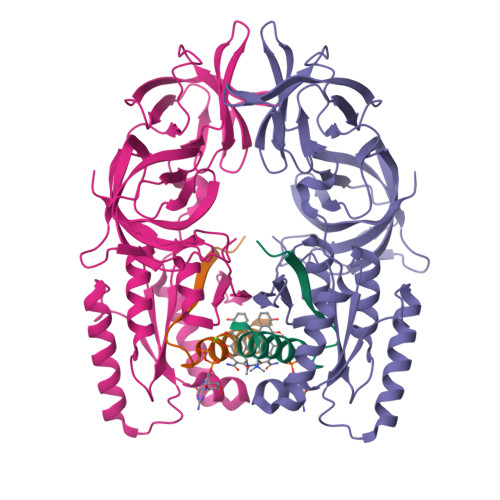
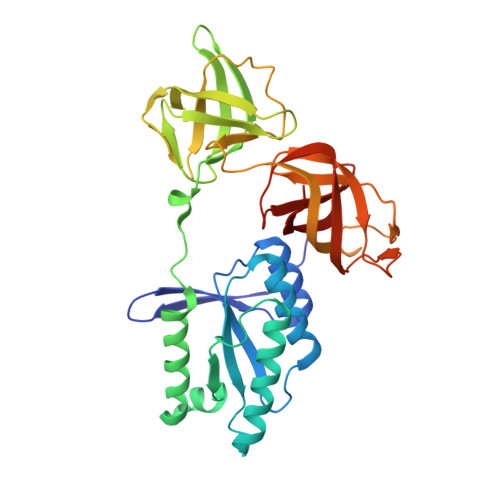
Molecular complementarity between tetracycline and the GTPase active site of elongation factor Tu.
Heffron, S.E., Mui, S., Aorora, A., Abel, K., Bergmann, E., Jurnak, F.(2006) Acta Crystallogr D Biol Crystallogr 62: 1392-1400
- PubMed: 17057344
- DOI: https://doi.org/10.1107/S0907444906035426
- Primary Citation of Related Structures:
2HCJ, 2HDN - PubMed Abstract:
Two crystal forms of a complex between trypsin-modified elongation factor Tu-MgGDP from Escherichia coli and the antibiotic tetracycline have been solved by X-ray diffraction analysis to resolutions of 2.8 and 2.1 A, respectively. In the P2(1) form, cocrystals were grown from a solution mixture of the protein and tetracycline. Six copies of the trypsin-modified EF-Tu-MgGDP-tetracycline complex are arranged as three sets of dimers in the asymmetric unit. In the second crystal form, tetracycline was diffused into P4(3)2(1)2 crystals, resulting in a monomeric complex in the asymmetric unit. Atomic coordinates have been refined to crystallographic R factors of 18.0% for the P2(1) form and 20.0% for the P4(3)2(1)2 form. In both complexes, tetracycline makes significant interactions with the GTPase active site of EF-Tu. The phenoldiketone moiety of tetracycline interacts directly with the Mg(2+), the alpha-phosphate group of GDP and two amino acids, Thr25 and Asp80, which are conserved in the GX(4)GKS/T and DX(2)G sequence motifs found in all GTPases and many ATPases. The molecular complementarity, previously unrecognized between invariant groups present in all GTPase/ATPases and the active moiety of tetracycline, may have wide-ranging implications for all drugs containing the phenoldiketone moiety as well as for the design of new compounds targeted against a broad range of GTPases or ATPases.
Organizational Affiliation:
Department of Physiology and Biophysics, 346-D Med Sci I, University of California, Irvine, CA 92697-4560, USA.