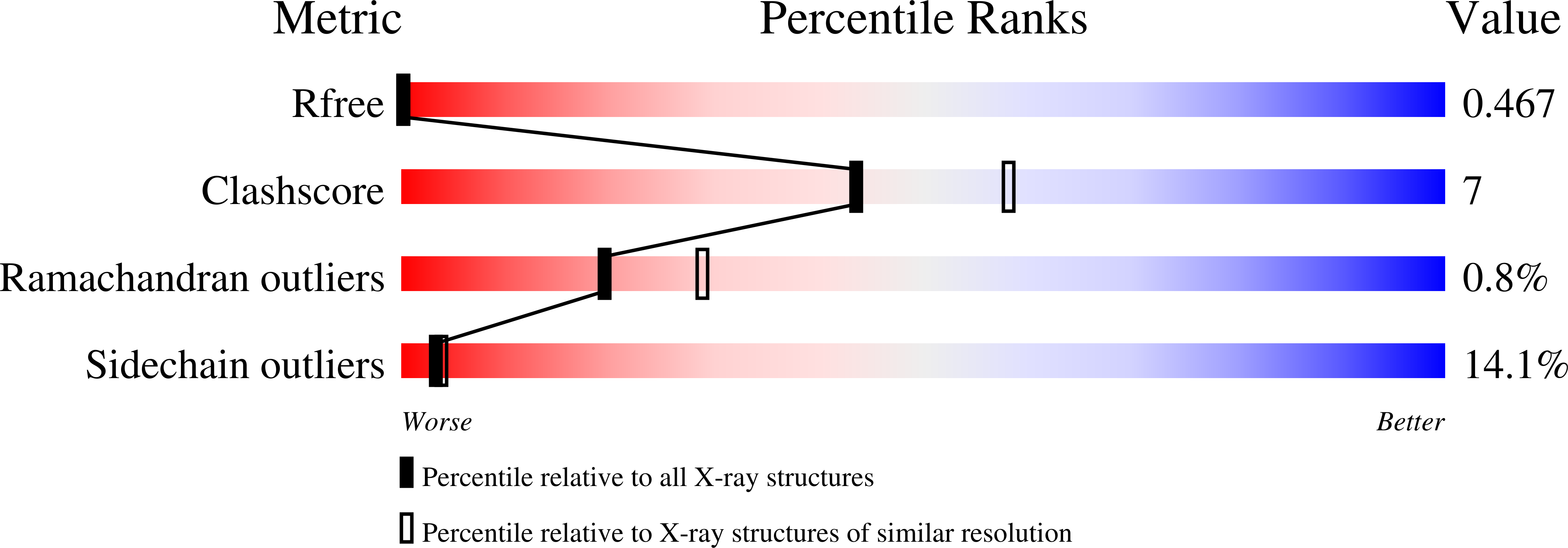
Structural Insights Into Fusidic Acid Resistance and Sensitivity in EF-G
Hansson, S., Singh, R., Gudkov, A.T., Liljas, A., Logan, D.T.(2005) J Mol Biol 348: 939
- PubMed: 15843024
- DOI: https://doi.org/10.1016/j.jmb.2005.02.066
- Primary Citation of Related Structures:
2BM0, 2BM1 - PubMed Abstract:
Fusidic acid (FA) is a steroid antibiotic commonly used against Gram positive bacterial infections. It inhibits protein synthesis by stalling elongation factor G (EF-G) on the ribosome after translocation. A significant number of the mutations conferring strong FA resistance have been mapped at the interfaces between domains G, III and V of EF-G. However, direct information on how such mutations affect the structure has hitherto not been available. Here we present the crystal structures of two mutants of Thermus thermophilus EF-G, G16V and T84A, which exhibit FA hypersensitivity and resistance in vitro, respectively. These mutants also have higher and lower affinity for GTP respectively than wild-type EF-G. The mutations cause significant conformational changes in the switch II loop that have opposite effects on the position of a key residue, Phe90, which undergoes large conformational changes. This correlates with the importance of Phe90 in FA sensitivity reported in previous studies. These structures substantiate the importance of the domain G/domain III/domain V interfaces as a key component of the FA binding site. The mutations also cause subtle changes in the environment of the "P-loop lysine", Lys25. This led us to examine the conformation of the equivalent residue in all structures of translational GTPases, which revealed that EF-G and eEF2 form a group separate from the others and suggested that the role of Lys25 may be different in the two groups.
Organizational Affiliation:
Department of Molecular Biophysics, Lund University, Box 124, S-221 00 Lund, Sweden.