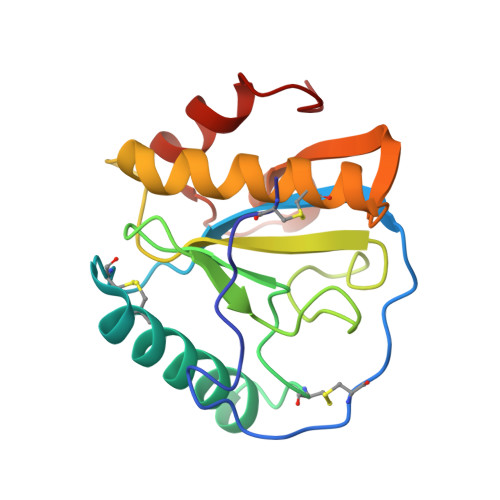
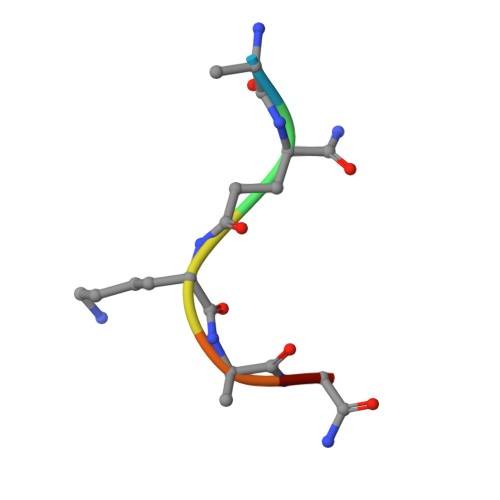
Crystal structure of human peptidoglycan recognition protein I alpha bound to a muramyl pentapeptide from Gram-positive bacteria.
Guan, R., Brown, P.H., Swaminathan, C.P., Roychowdhury, A., Boons, G.J., Mariuzza, R.A.(2006) Protein Sci 15: 1199-1206
- PubMed: 16641493
- DOI: https://doi.org/10.1110/ps.062077606
- Primary Citation of Related Structures:
2APH - PubMed Abstract:
Peptidoglycan recognition proteins (PGRPs) are pattern recognition receptors of the innate immune system that bind bacterial peptidoglycans (PGNs). We determined the crystal structure, to 2.1 A resolution, of the C-terminal PGN-binding domain of human PGRP-I alpha in complex with a muramyl pentapeptide (MPP) from Gram-positive bacteria containing a complete peptide stem (L-Ala-D-isoGln-L-Lys-D-Ala-D-Ala). The structure reveals important features not observed previously in the complex between PGRP-I alpha and a muramyl tripeptide lacking D-Ala at stem positions 4 and 5. Most notable are ligand-induced structural rearrangements in the PGN-binding site that are essential for entry of the C-terminal portion of the peptide stem and for locking MPP in the binding groove. We propose that similar structural rearrangements to accommodate the PGN stem likely characterize many PGRPs, both mammalian and insect.
Organizational Affiliation:
Center for Advanced Research in Biotechnology, W.M. Keck Laboratory for Structural Biology, University of Maryland Biotechnology Institute, Rockville, Maryland 20850, USA.