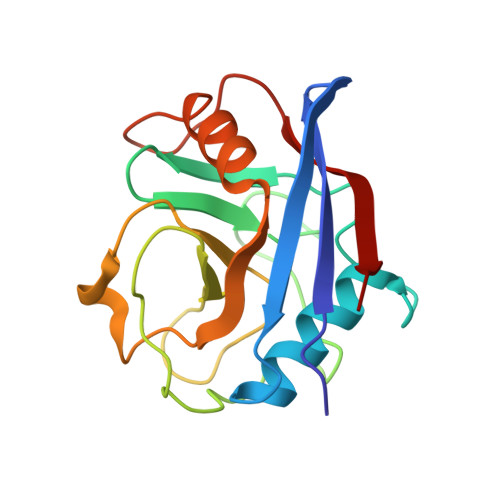
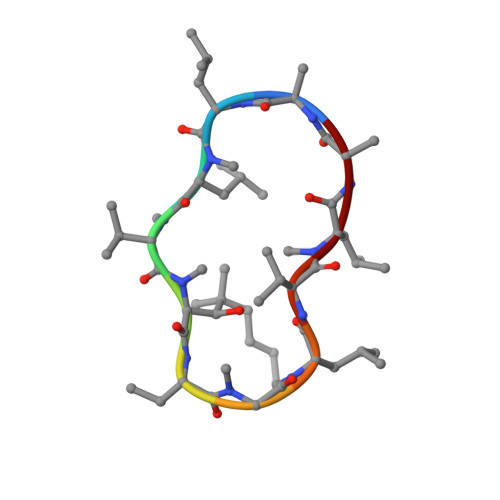
Crystal Structures of Cyclophilin a Complexed with Cyclosporin a and N-Methyl-4-[(E)-2-Butenyl]-4,4-Dimethylthreonine Cyclosporin A.
Ke, H., Mayrose, D., Belshaw, P.J., Alberg, D.G., Schreiber, S.L., Chang, Z.Y., Etzkorn, F.A., Ho, S., Walsh, C.T.(1994) Structure 2: 33
- PubMed: 8075981
- DOI: https://doi.org/10.1016/s0969-2126(00)00006-x
- Primary Citation of Related Structures:
2RMA, 2RMB - PubMed Abstract:
Cyclophilin (CyP) is a ubiquitious intracellular protein that binds the immunosuppressive drug cyclosporin A (CsA). CyP-CsA forms a ternary complex with calcineurin and thereby inhibits T-cell activation. CyP also has enzymatic activity, catalyzing the cis-trans isomerization of peptidyl-prolyl amide bonds. We have determined the structure of human cyclophilin A (CyPA) complexed with CsA to 2.1 A resolution. We also report here the structure of CyPA complexed with an analog of CsA, CsA (MeBm2t1-CsA), which binds less well to CyPA, but has increased immunosuppressive activity. Comparison of these structures with previously determined structures of unligated CyPA and CyPA complexed with a candidate substrate for the isomerase activity, the dipeptide AlaPro, reveals that subtle conformational changes occur in both CsA and CyPA on complex formation. MeBm2t1-CsA binds to CyPA in an essentially similar manner to CsA. The 100-fold weaker affinity of its binding may be attributable to the close contact between MeBmt1 and the active site residue Ala103 of CyPA, which causes small conformational changes in both protein and drug. One change, the slight movement of MeLeu6 in CsA relative to MeBm2t1-CsA, may be at least partially responsible for the higher affinity of the CyPA-MeBm2t1-CsA complex for calcineurin. Our comparison between CyPA-CsA and CyPA-AlaPro suggests that CsA is probably not an analog of the natural substrate, confirming that the catalytic activity of CyPA is not related to its role in immunosuppression either structurally or functionally.
Organizational Affiliation:
Department of Biochemistry and Biophysics, School of Medicine, University of North Carolina, Chapel HIll 27599.