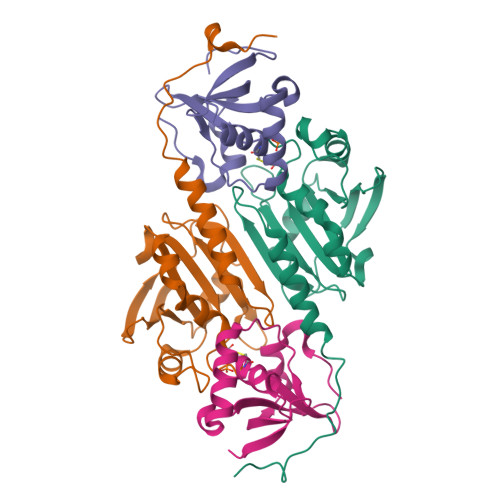
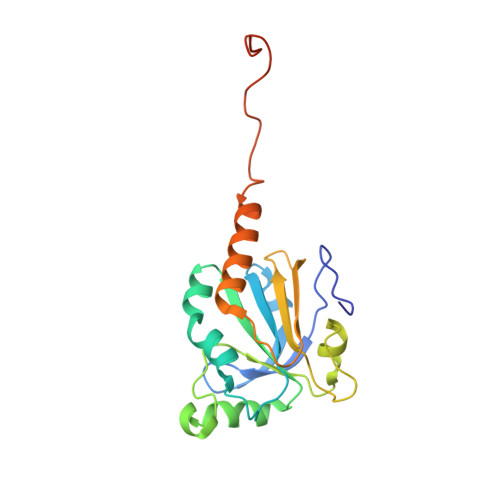
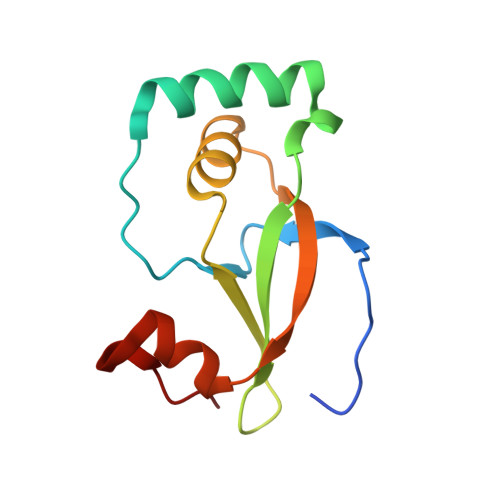
Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace.
Jonsson, T.J., Johnson, L.C., Lowther, W.T.(2008) Nature 451: 98-101
- PubMed: 18172504
- DOI: https://doi.org/10.1038/nature06415
- Primary Citation of Related Structures:
2RII - PubMed Abstract:
Typical 2-Cys peroxiredoxins (Prxs) have an important role in regulating hydrogen peroxide-mediated cell signalling. In this process, Prxs can become inactivated through the hyperoxidation of an active site Cys residue to Cys sulphinic acid. The unique repair of this moiety by sulphiredoxin (Srx) restores peroxidase activity and terminates the signal. The hyperoxidized form of Prx exists as a stable decameric structure with each active site buried. Therefore, it is unclear how Srx can access the sulphinic acid moiety. Here we present the 2.6 A crystal structure of the human Srx-PrxI complex. This complex reveals the complete unfolding of the carboxy terminus of Prx, and its unexpected packing onto the backside of Srx away from the Srx active site. Binding studies and activity analyses of site-directed mutants at this interface show that the interaction is required for repair to occur. Moreover, rearrangements in the Prx active site lead to a juxtaposition of the Prx Gly-Gly-Leu-Gly and Srx ATP-binding motifs, providing a structural basis for the first step of the catalytic mechanism. The results also suggest that the observed interactions may represent a common mode for other proteins to bind to Prxs.
Organizational Affiliation:
Center for Structural Biology and Department of Biochemistry, Wake Forest University School of Medicine, Medical Center Boulevard, Winston-Salem, North Carolina 27157, USA.