Binding of Rac1, Rnd1, and RhoD to a novel Rho GTPase interaction motif destabilizes dimerization of the plexin-B1 effector domain.
Tong, Y., Chugha, P., Hota, P.K., Alviani, R.S., Li, M., Tempel, W., Shen, L., Park, H.W., Buck, M.(2007) J Biol Chem 282: 37215-37224
- PubMed: 17916560
- DOI: https://doi.org/10.1074/jbc.M703800200
- Primary Citation of Related Structures:
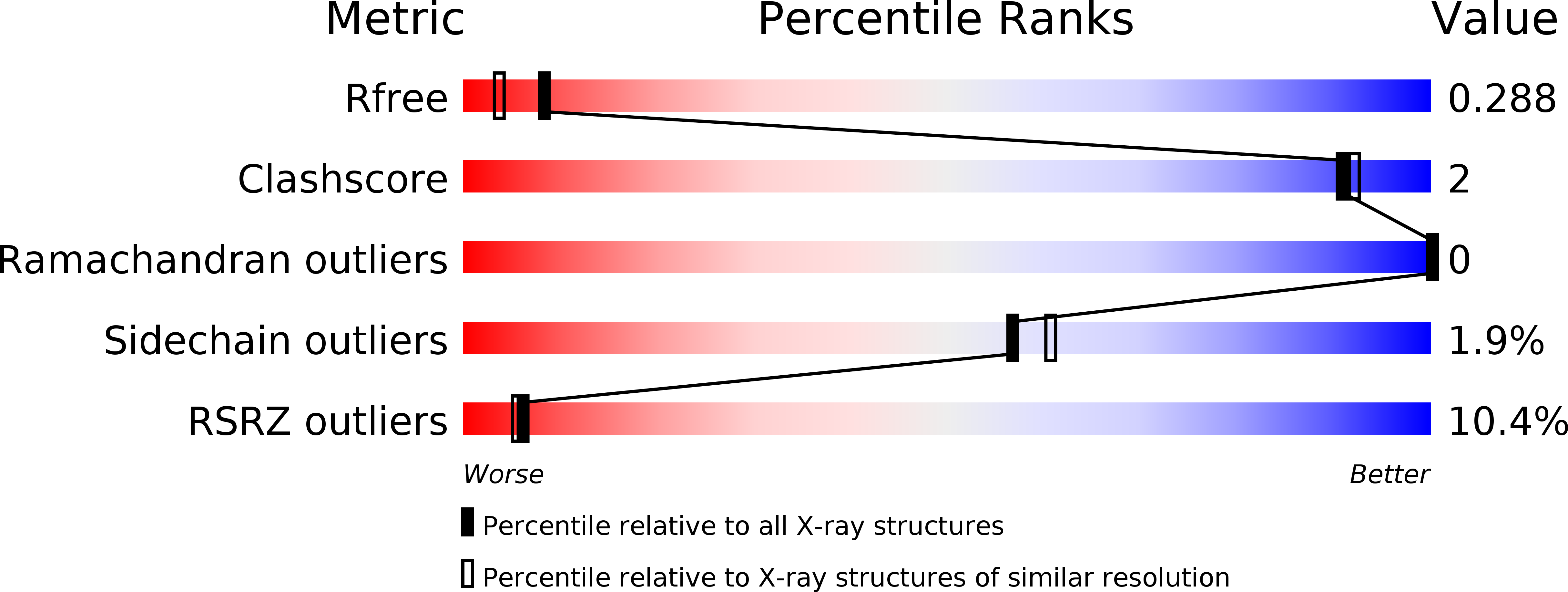
2R2O - PubMed Abstract:
Plexins are the first known transmembrane receptors that interact directly with small GTPases. On binding to certain Rho family GTPases, the receptor regulates the remodeling of the actin cytoskeleton and alters cell movement in response to semaphorin guidance cues. In a joint solution NMR spectroscopy and x-ray crystallographic study, we characterize a 120-residue cytoplasmic independent folding domain of plexin-B1 that directly binds three Rho family GTPases, Rac1, Rnd1, and RhoD. The NMR data show that, surprisingly, the Cdc42/Rac interactive binding-like motif of plexin-B1 is not involved in this interaction. Instead, all three GTPases interact with the same region, beta-strands 3 and 4 and a short alpha-helical segment of the plexin domain. The 2.0 A resolution x-ray structure shows that these segments are brought together by the tertiary structure of the ubiquitin-like fold. In the crystal, the protein is dimerized with C2 symmetry through a four-stranded antiparallel beta-sheet that is formed outside the fold by a long loop between the monomers. This region is adjacent to the GTPase binding motifs identified by NMR. Destabilization of the dimer in solution by binding of any one of the three GTPases suggests a model for receptor regulation that involves bidirectional signaling. The model implies a multifunctional role for the GTPase-plexin interaction that includes conformational change and a localization of active receptors in the signaling mechanism.
Organizational Affiliation:
Department of Physiology, Case Western Reserve University School of Medicine, Cleveland, Ohio 44106, USA.