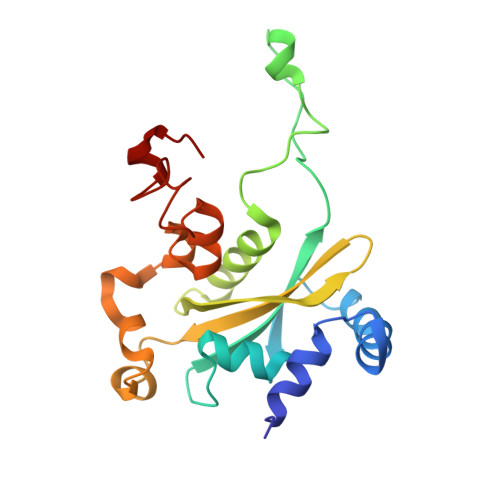
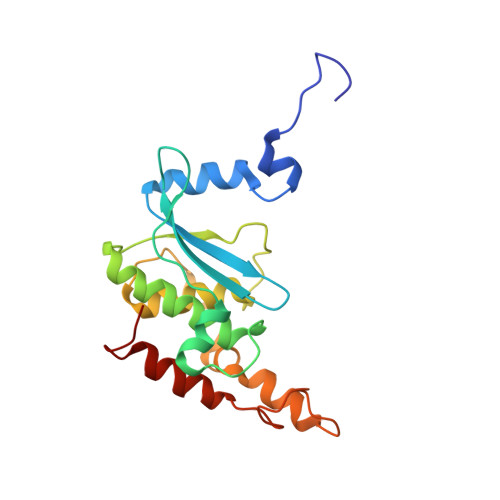
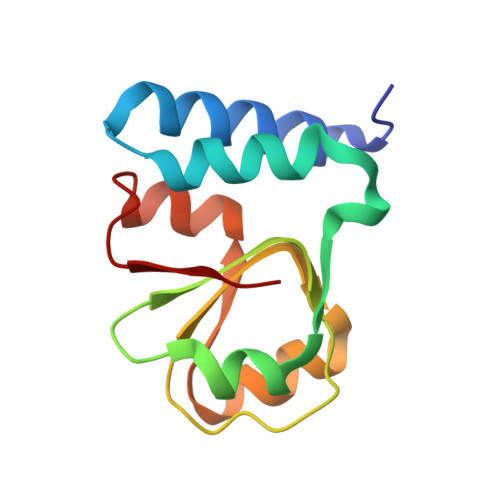
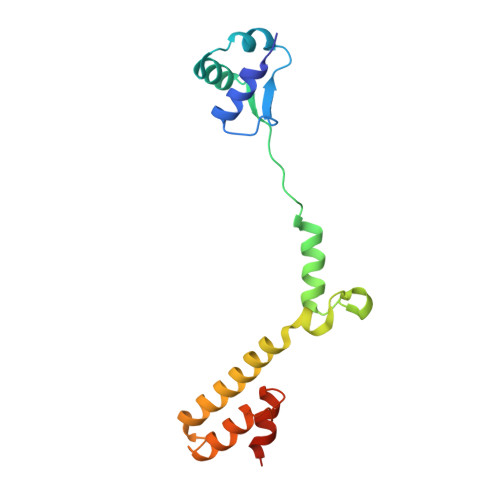
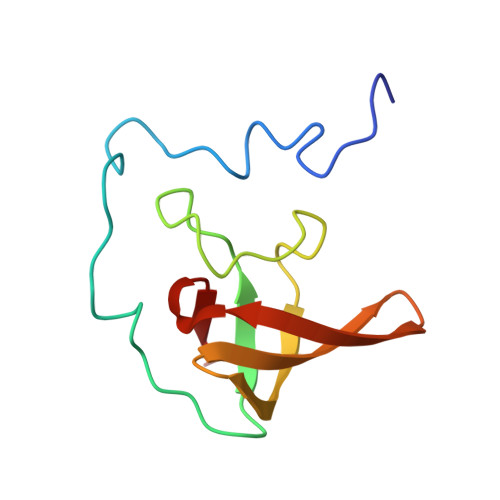
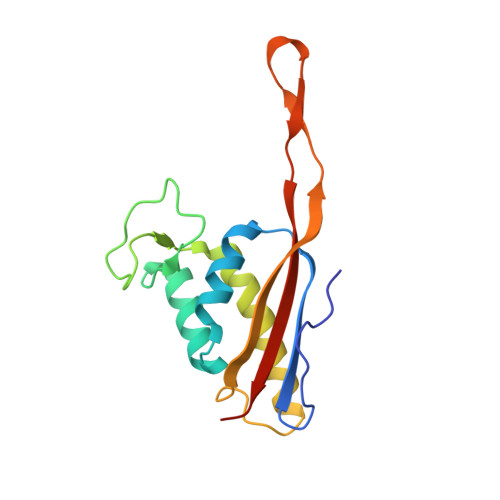
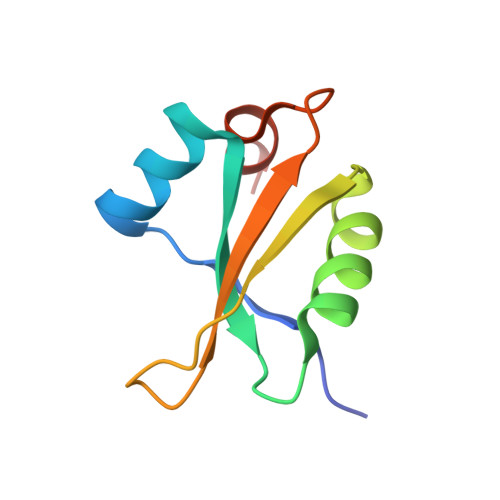
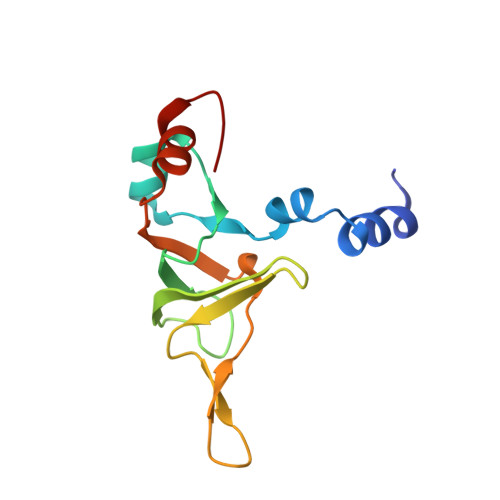
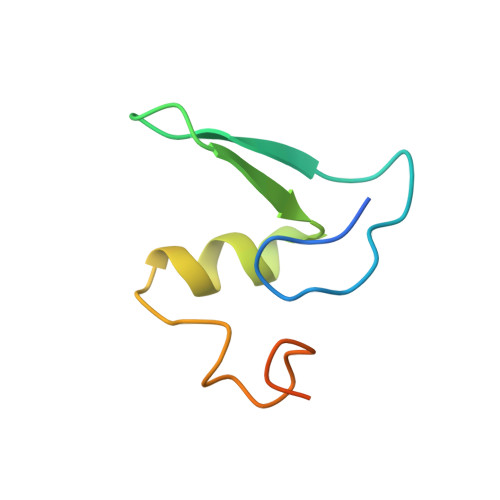
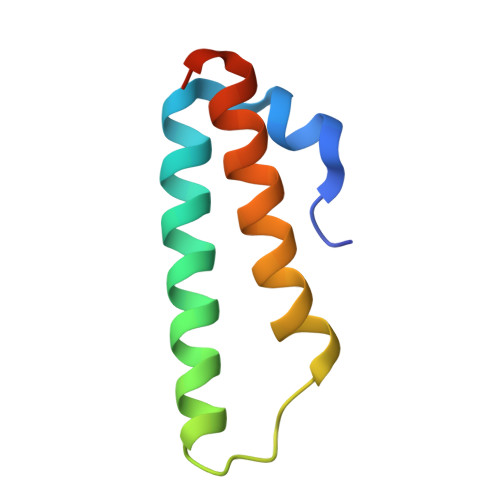
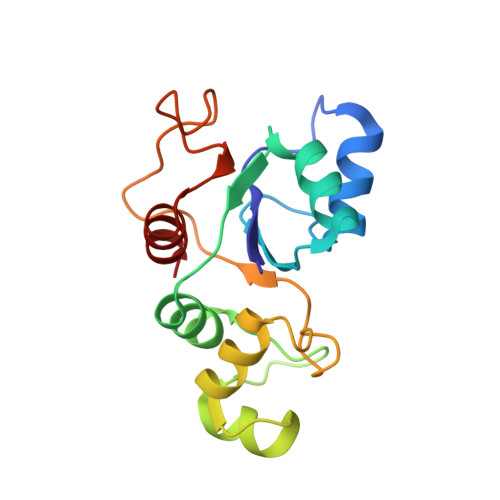
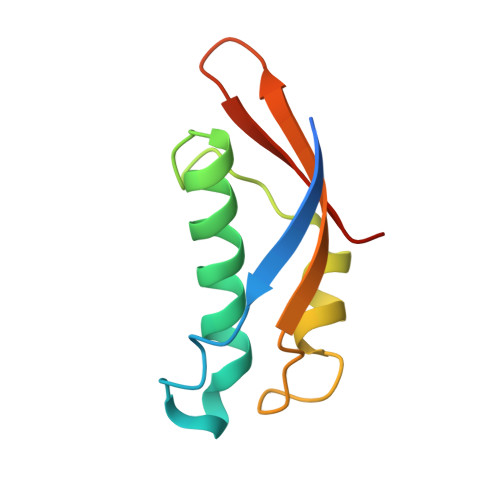
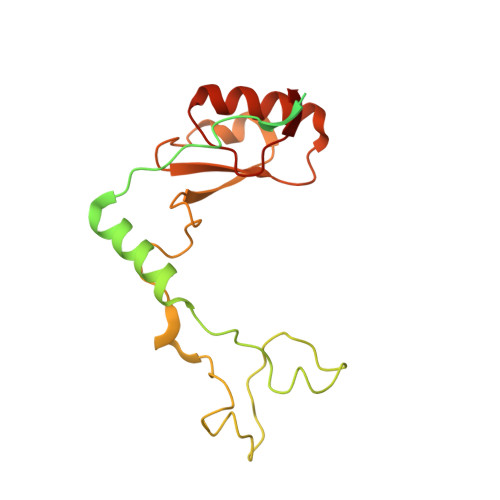
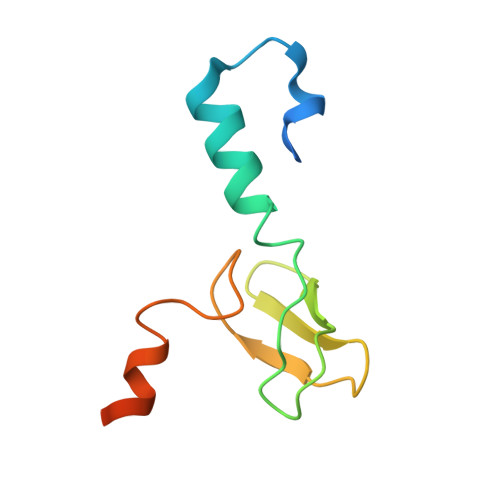
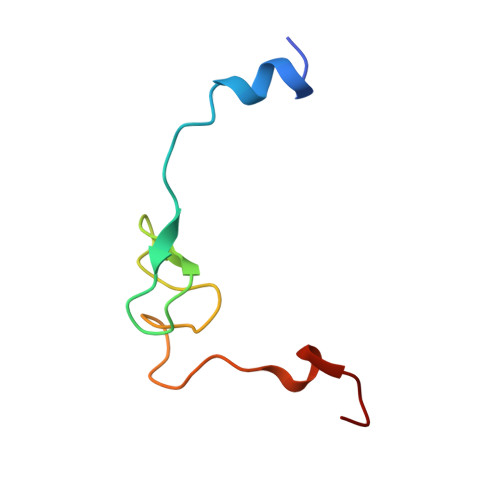
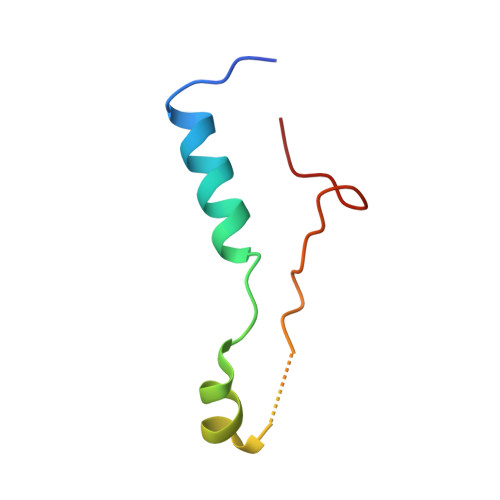
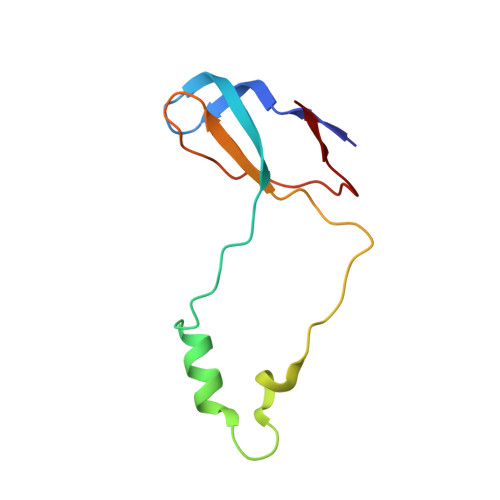
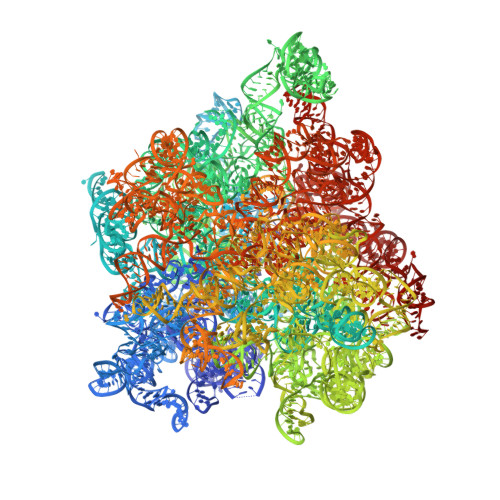
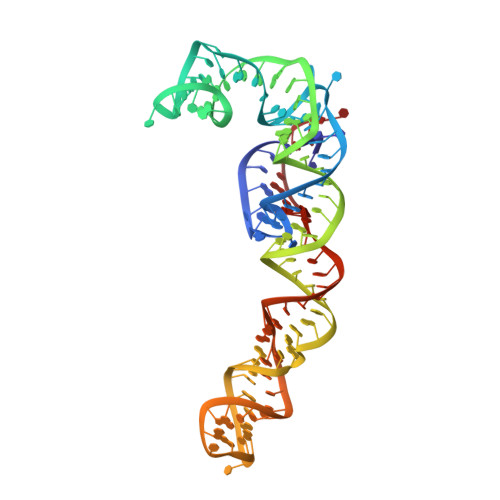
Structure of the base of the L7/L12 stalk of the Haloarcula marismortui large ribosomal subunit: Analysis of L11 movements
Kavran, J.M., Steitz, T.A.(2007) J Mol Biol 371: 1047-1059
- PubMed: 17599351
- DOI: https://doi.org/10.1016/j.jmb.2007.05.091
- Primary Citation of Related Structures:
2QA4 - PubMed Abstract:
Initiation factors, elongation factors, and release factors all interact with the L7/L12 stalk of the large ribosomal subunit during their respective GTP-dependent cycles on the ribosome. Electron density corresponding to the stalk is not present in previous crystal structures of either 50 S subunits or 70 S ribosomes. We have now discovered conditions that result in a more ordered factor-binding center in the Haloarcula marismortui (H.ma) large ribosomal subunit crystals and consequently allows the visualization of the full-length L11, the N-terminal domain (NTD) of L10 and helices 43 and 44 of 23 S rRNA. The resulting model is currently the most complete reported structure of a L7/L12 stalk in the context of a ribosome. This region contains a series of intermolecular interfaces that are smaller than those typically seen in other ribonucleoprotein interactions within the 50 S subunit. Comparisons of the L11 NTD position between the current structure, which is has an NTD splayed out with respect to previous structures, and other structures of ribosomes in different functional states demonstrates a dynamic range of L11 NTD movements. We propose that the L11 NTD moves through three different relative positions during the translational cycle: apo-ribosome, factor-bound pre-GTP hydrolysis and post-GTP hydrolysis. These positions outline a pathway for L11 NTD movements that are dependent on the specific nucleotide state of the bound ligand. These three states are represented by the orientations of the L11 NTD relative to the ribosome and suggest that L11 may play a more specialized role in the factor binding cycle than previously appreciated.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.