Purification, crystallization and preliminary crystallographic analysis of deoxyuridine triphosphate nucleotidohydrolase from Arabidopsis thaliana.
Bajaj, M., Moriyama, H.(2007) Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 409-411
- PubMed: 17565183
- DOI: https://doi.org/10.1107/S1744309107016004
- Primary Citation of Related Structures:
2PC5 - PubMed Abstract:
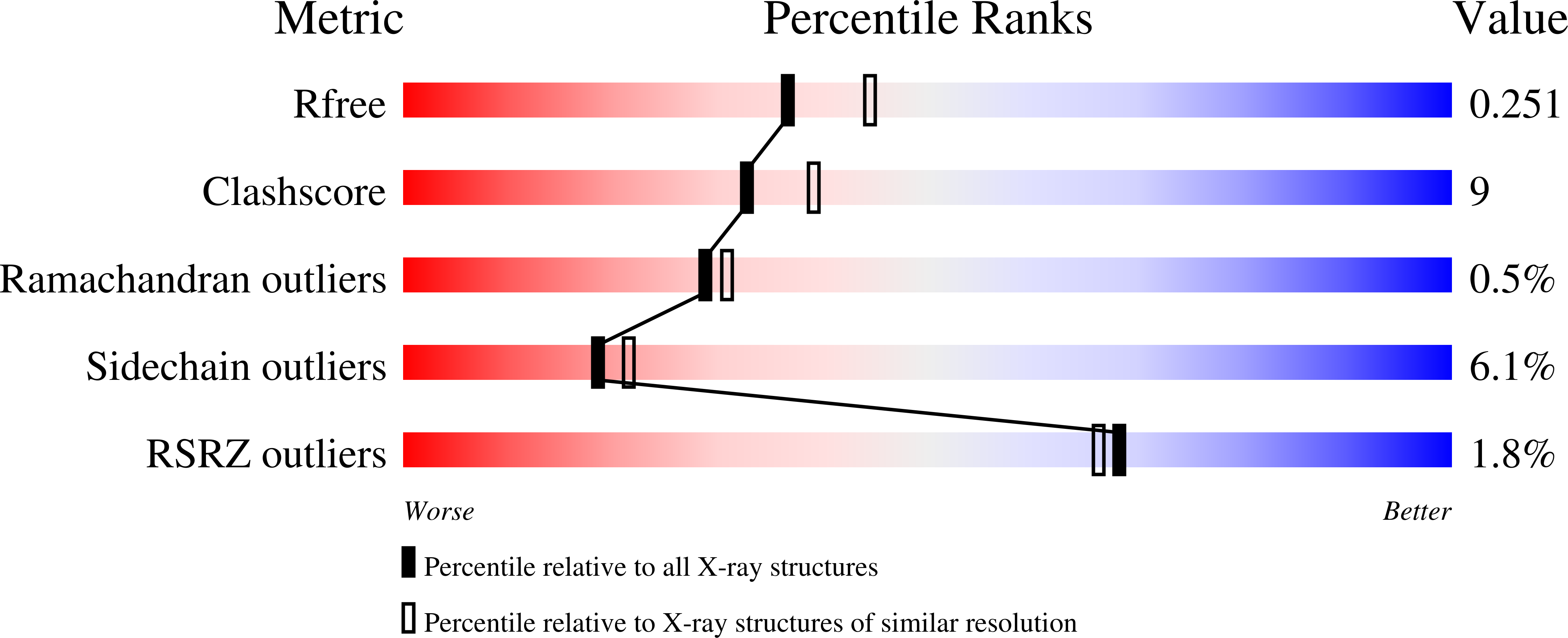
The deoxyuridine triphosphate nucleotidohydrolase gene from Arabidopsis thaliana was expressed and the gene product was purified. Crystallization was performed by the hanging-drop vapour-diffusion method at 298 K using 2 M ammonium sulfate as the precipitant. X-ray diffraction data were collected to 2.2 A resolution using Cu K alpha radiation. The crystal belongs to the orthorhombic space group P2(1)2(1)2(1), with unit-cell parameters a = 69.90, b = 70.86 A, c = 75.55 A. Assuming the presence of a trimer in the asymmetric unit, the solvent content was 30%, with a V(M) of 1.8 A3 Da(-1).
Organizational Affiliation:
School of Biological Sciences, University of Nebraska-Lincoln, Manter Hall, Lincoln, Nebraska 68588-0304, USA.