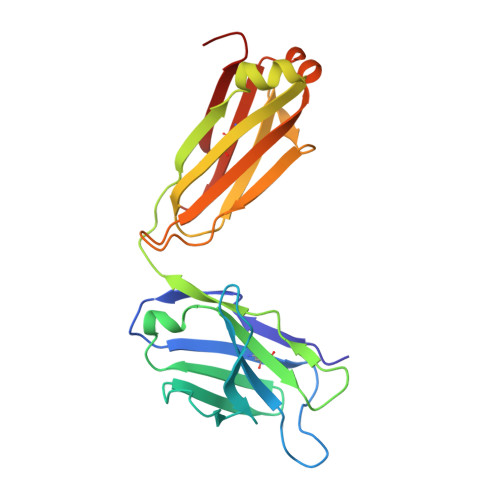
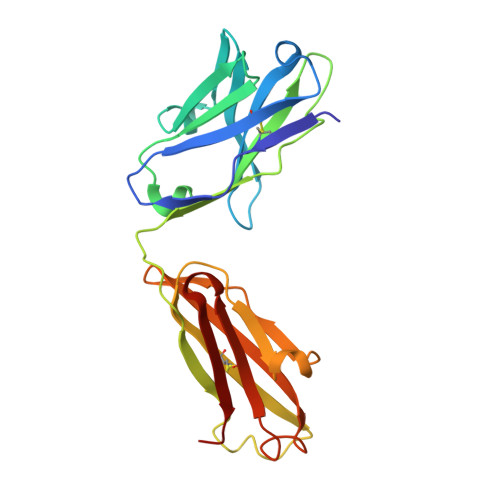
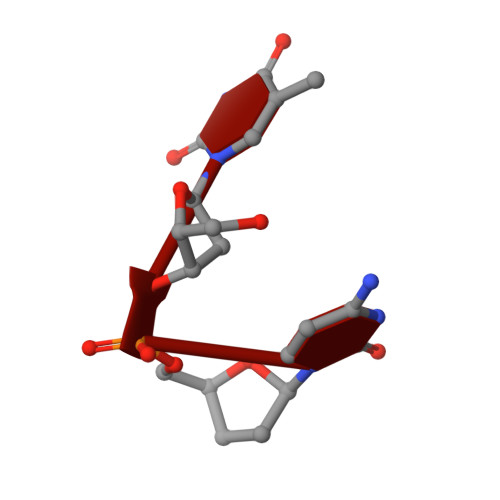
Specific recognition of a DNA immunogen by its elicited antibody
Sanguineti, S., Centeno Crowley, J.M., Lodeiro Merlo, M.F., Cerutti, M.L., Wilson, I.A., Goldbaum, F.A., Stanfield, R.L., de Prat-Gay, G.(2007) J Mol Biol 370: 183-195
- PubMed: 17512945
- DOI: https://doi.org/10.1016/j.jmb.2007.04.046
- Primary Citation of Related Structures:
2OJZ, 2OK0 - PubMed Abstract:
DNA recognition by antibodies is a key feature of autoimmune diseases, yet model systems with structural information are very limited. The monoclonal antibody ED-10 recognizes one of the strands of the DNA duplex used in the immunogenic complex. Modifications of the 5' end decrease the binding affinity and short oligonucleotides retain high binding affinity. We determined crystal structures for the Fab bound to a 6-mer oligonucleotide containing the specific sequence that raised the antibody and compared it with the unliganded Fab. Only the first two bases from the 5' end (dTdC) display electron density and we observe four key hydrogen bonds at the interface. The thymine ring is stacked between TrpH50 and TrpH95, and the cytosine ring is packed against TyrL32. Upon DNA binding, TyrH97 and TrpH95 rearrange to allow subnanomolar binding affinity, five orders of magnitude higher than other reported complexes, possibly because of having gone through affinity maturation. This structure represents the first bona fide antibody DNA immunogen complex described in atomic detail.
Organizational Affiliation:
Instituto Leloir and CONICET, Patricias Argentinas 435, 1405 Buenos Aires, Argentina.