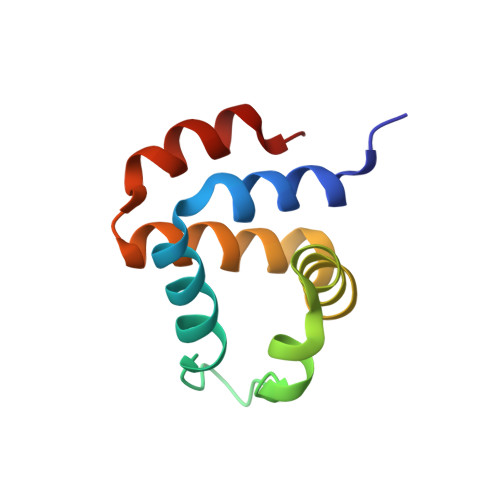
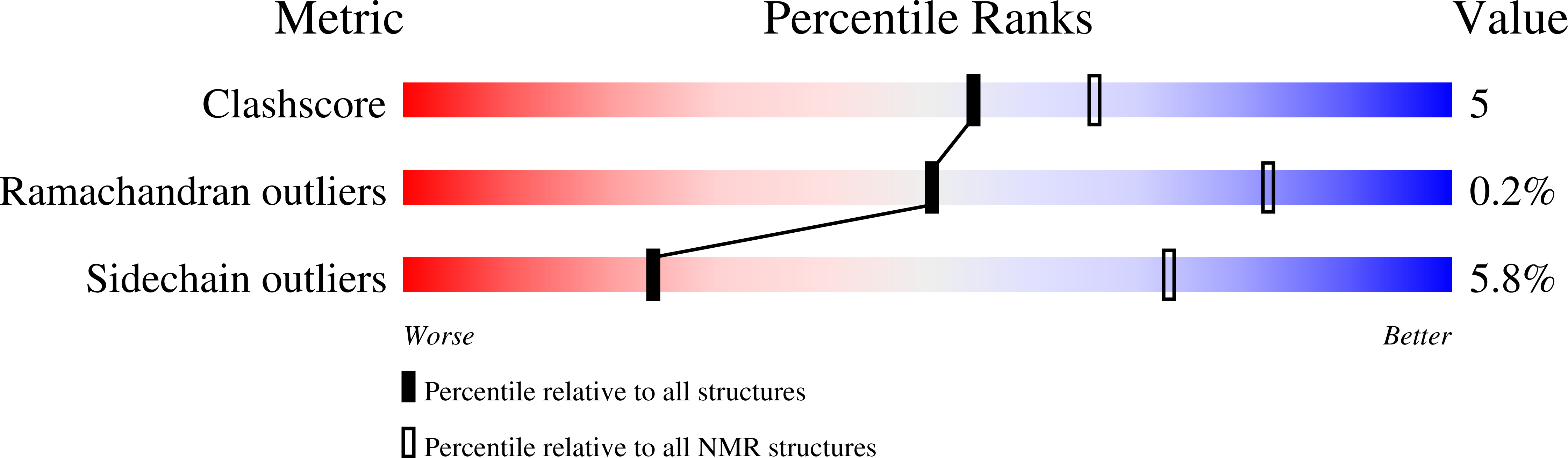ASC Pyrin Domain Self-associates and Binds NLRP3 Protein Using Equivalent Binding Interfaces.
Oroz, J., Barrera-Vilarmau, S., Alfonso, C., Rivas, G., de Alba, E.(2016) J Biol Chem 291: 19487-19501
- PubMed: 27432880
- DOI: https://doi.org/10.1074/jbc.M116.741082
- Primary Citation of Related Structures:
2NAQ - PubMed Abstract:
Death domain superfamily members typically act as adaptors mediating in the assembly of supramolecular complexes with critical apoptosis and inflammation functions. These modular proteins consist of death domains, death effector domains, caspase recruitment domains, and pyrin domains (PYD). Despite the high structural similarity among them, only homotypic interactions participate in complex formation, suggesting that subtle factors differentiate each interaction type. It is thus critical to identify these factors as an essential step toward the understanding of the molecular basis of apoptosis and inflammation. The proteins apoptosis-associated speck-like protein containing a CARD (ASC) and NLRP3 play key roles in the regulation of apoptosis and inflammation through self-association and protein-protein interactions mediated by their PYDs. To better understand the molecular basis of their function, we have characterized ASC and NLRP3 PYD self-association and their intermolecular interaction by solution NMR spectroscopy and analytical ultracentrifugation. We found that ASC self-associates and binds NLRP3 PYD through equivalent protein regions, with higher binding affinity for the latter. These regions are located at opposite sides of the protein allowing multimeric complex formation previously shown in ASC PYD fibril assemblies. We show that NLRP3 PYD coexists in solution as a monomer and highly populated large-order oligomerized species. Despite this, we determined its monomeric three-dimensional solution structure by NMR and characterized its binding to ASC PYD. Using our novel structural data, we propose molecular models of ASC·ASC and ASC·NLRP3 PYD early supramolecular complexes, providing new insights into the molecular mechanisms of inflammasome and apoptosis signaling.
Organizational Affiliation:
From the Centro de Investigaciones Biológicas, Departments of Chemical and Physical Biology and the German Center for Neurodegenerative Diseases (DZNE), ℅Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, Göttingen-37077, Germany, and jaor@nmr.mpibpc.mpg.de.