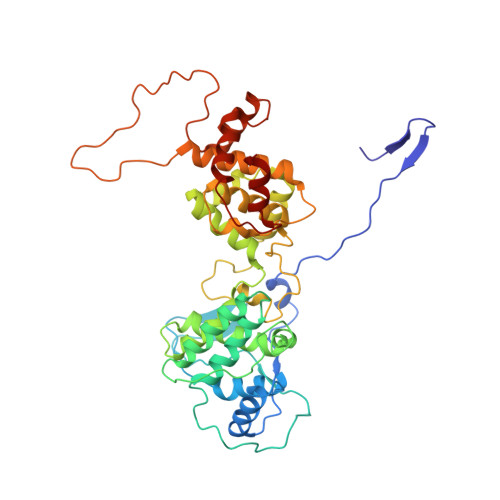
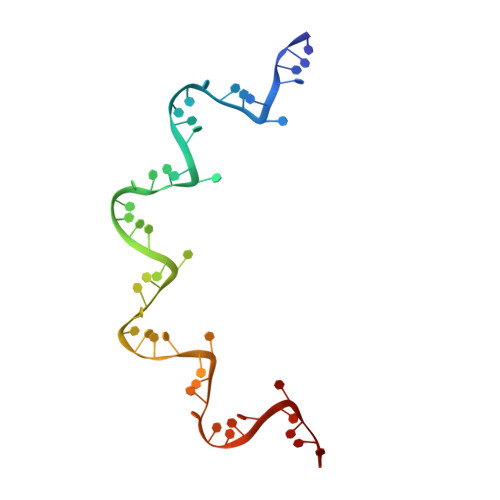Structure of the vesicular stomatitis virus nucleoprotein-RNA complex unveils how the RNA is sequestered
Green, T.J., Zhang, X., Wertz, G.W., Luo, M.(2006) Science 313: 357-360
- PubMed: 16778022
- DOI: https://doi.org/10.1126/science.1126953
- Primary Citation of Related Structures:
2GIC - PubMed Abstract:
Vesicular stomatitis virus is a negative-stranded RNA virus. Its nucleoprotein (N) binds the viral genomic RNA and is involved in multiple functions including transcription, replication, and assembly. We have determined a 2.9 angstrom structure of a complex containing 10 molecules of the N protein and 90 bases of RNA. The RNA is tightly sequestered in a cavity at the interface between two lobes of the N protein. This serves to protect the RNA in the absence of polynucleotide synthesis. For the RNA to be accessed, some conformational change in the N protein should be necessary.
Organizational Affiliation:
Department of Microbiology, School of Medicine, University of Alabama at Birmingham, 1025 18th Street South, Birmingham, AL 35294, USA.