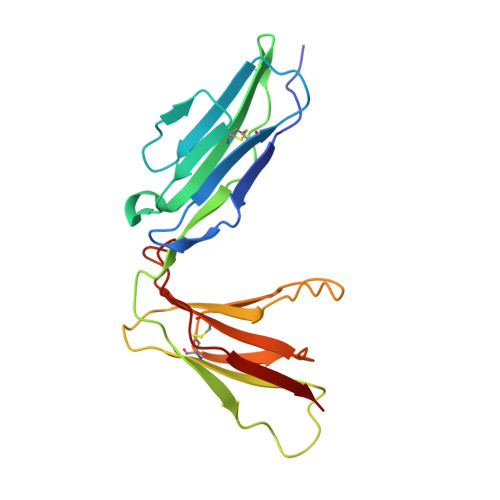
Structural basis for platelet collagen responses by the immune-type receptor glycoprotein VI.
Horii, K., Kahn, M.L., Herr, A.B.(2006) Blood 108: 936-942
- PubMed: 16861347
- DOI: https://doi.org/10.1182/blood-2006-01-010215
- Primary Citation of Related Structures:
2GI7 - PubMed Abstract:
Activation of circulating platelets by exposed vessel wall collagen is a primary step in the pathogenesis of heart attack and stroke, and drugs to block platelet activation have successfully reduced cardiovascular morbidity and mortality. In humans and mice, collagen activation of platelets is mediated by glycoprotein VI (GPVI), a receptor that is homologous to immune receptors but bears little sequence similarity to known matrix protein adhesion receptors. Here we present the crystal structure of the collagen-binding domain of human GPVI and characterize its interaction with a collagen-related peptide. Like related immune receptors, GPVI contains 2 immunoglobulin-like domains arranged in a perpendicular orientation. Significantly, GPVI forms a back-to-back dimer in the crystal, an arrangement that could explain data previously obtained from cell-surface GPVI inhibition studies. Docking algorithms identify 2 parallel grooves on the GPVI dimer surface as collagen-binding sites, and the orientation and spacing of these grooves precisely match the dimensions of an intact collagen fiber. These findings provide a structural basis for the ability of an immune-type receptor to generate signaling responses to collagen and for the development of GPVI inhibitors as new therapies for human cardiovascular disease.
Organizational Affiliation:
Department of Molecular Genetics, Biochemistry & Microbiology, University of Cincinnati College of Medicine, 231 Albert Sabin Way, OH 45267-0524, USA.