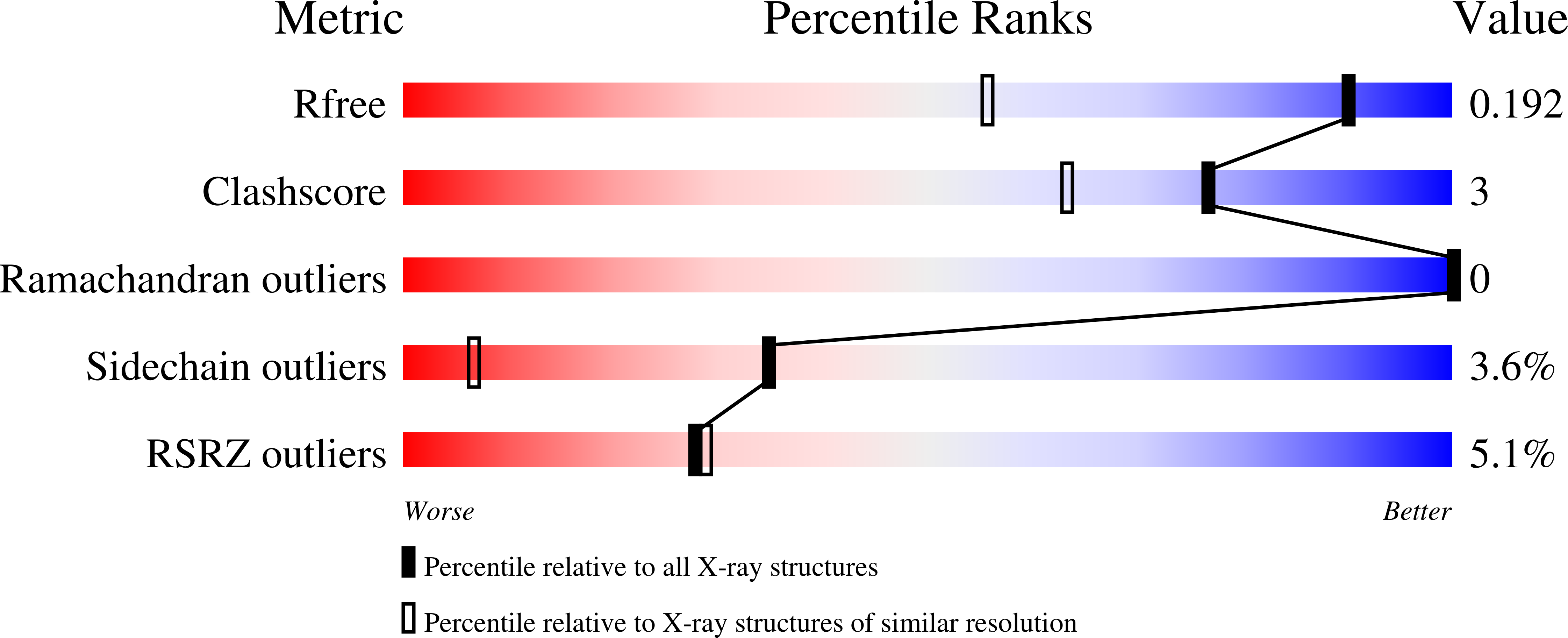
The 1.38 A crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway.
Qiu, Y., Zhang, R., Binkowski, T.A., Tereshko, V., Joachimiak, A., Kossiakoff, A.(2008) Proteins 71: 525-533
- PubMed: 18175314
- DOI: https://doi.org/10.1002/prot.21828
- Primary Citation of Related Structures:
1S9U - PubMed Abstract:
The DmsD protein is necessary for the biogenesis of dimethyl sulphoxide (DMSO) reductase in many prokaryotes. It performs a critical chaperone function initiated through its binding to the twin-arginine signal peptide of DmsA, the catalytic subunit of DMSO reductase. Upon binding to DmsD, DmsA is translocated to the periplasm via the so-called twin-arginine translocation (Tat) pathway. Here we report the 1.38 A crystal structure of the protein DmsD from Salmonella typhimurium and compare it with a close functional homolog, TorD. DmsD has an all-alpha fold structure with a notable helical extension located at its N-terminus with two solvent exposed hydrophobic residues. A major difference between DmsD and TorD is that TorD structure is a domain-swapped dimer, while DmsD exists as a monomer. Nevertheless, these two proteins have a number of common features suggesting they function by using similar mechanisms. A possible signal peptide-binding site is proposed based on structural similarities. Computational analysis was used to identify a potential GTP binding pocket on similar surfaces of DmsD and TorD structures.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, Illinois 60637, USA.