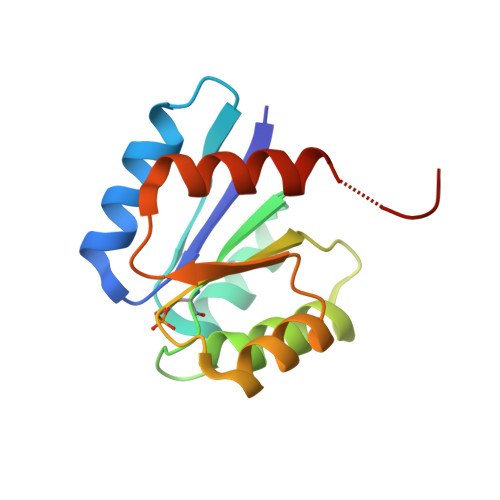
Phosphorylated aspartate in the structure of a response regulator protein.
Lewis, R.J., Brannigan, J.A., Muchova, K., Barak, I., Wilkinson, A.J.(1999) J Mol Biol 294: 9-15
- PubMed: 10556024
- DOI: https://doi.org/10.1006/jmbi.1999.3261
- Primary Citation of Related Structures:
1QMP - PubMed Abstract:
Phosphorylation of aspartic acid residues is the hallmark of two- component signal transduction systems that orchestrate the adaptive responses of micro-organisms to changes in their surroundings. Two-component systems consist of a sensor kinase that interprets environmental signals and a response regulator that activates the appropriate physiological response. Although structures of response regulators are known, little is understood about their activated phosphorylated forms, due to the intrinsic instability of the acid phosphate linkage. Here, we report the phosphorylated structure of the receiver/phosphoacceptor domain of Spo0A, the master regulator of sporulation, from Bacillus stearothermophilus. The phosphoryl group is covalently bonded to the invariant aspartate 55, and co-ordinated to a nearby divalent metal cation, with both species fulfilling their electrostatic potential through interactions with solvent water molecules, the protein main chain, and with side-chains of amino acid residues strongly conserved across the response regulator family. This is the first direct visualisation of a phosphoryl group covalently linked to an aspartic acid residue in any protein, with implications for signalling within the response regulator family.
Organizational Affiliation:
Structural Biology Laboratory, Department of Chemistry, University of York, York, YO10 5DD, UK.