Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
Kwon, A.R., Kessler, B.M., Overkleeft, H.S., Mckay, D.B.(2003) J Mol Biology 330: 185
- PubMed: 12823960
- DOI: https://doi.org/10.1016/s0022-2836(03)00580-1
- Primary Citation of Related Structures:
1OFH, 1OFI - PubMed Abstract:
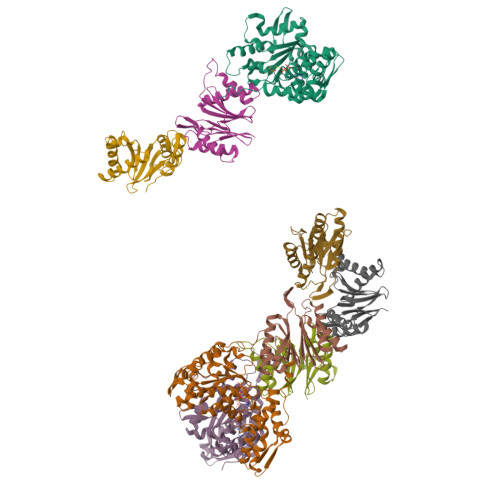
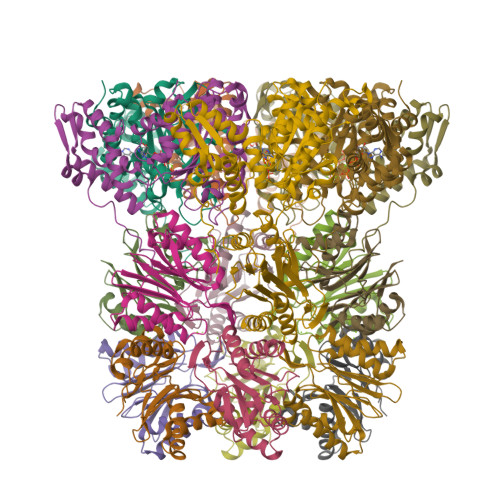
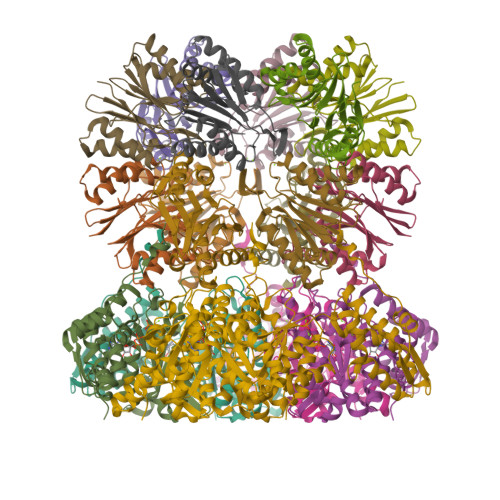
In the prokaryotic homolog of the eukaryotic proteasome, HslUV, the "double donut" HslV protease is allosterically activated by HslU, an AAA protein of the Clp/Hsp100 family consisting of three (amino-terminal, carboxy-terminal, and intermediate) domains. The intermediate domains of HslU, which extend like tentacles from the hexameric ring formed by the amino-terminal and carboxy-terminal domains, have been deleted; an asymmetric HslU(DeltaI)(6)HslV(12) complex has been crystallized; and the structure has been solved to 2.5A resolution, revealing an assembly in which a HslU(DeltaI) hexamer binds one end of the HslV dodecamer. The conformation of the protomers of the HslU(DeltaI)-complexed HslV hexamer is similar to that in the symmetric wild-type HslUV complex, while the protomer conformation of the uncomplexed HslV hexamer is similar to that of HslV alone. Reaction in the crystals with a vinyl sulfone inhibitor reveals that the HslU(DeltaI)-complexed HslV hexamer is active, while the uncomplexed HslV hexamer is inactive. These results confirm that HslV can be activated by binding of a hexameric HslU(DeltaI)(6) ring lacking the I domains, that activation is effected through a conformational change in HslV rather than through alteration of the size of the entry channel into the protease catalytic cavity, and that the two HslV(6) rings in the protease dodecamer are activated independently rather than cooperatively.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine, Sherman Fairchild Building, Stanford, CA 94305, USA.