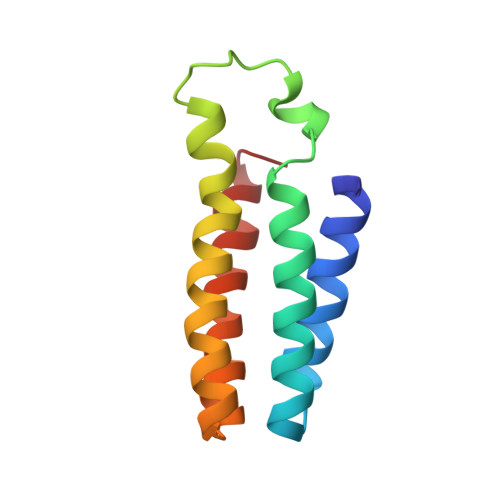
Structural consequences of b- to c-type heme conversion in oxidized Escherichia coli cytochrome b562.
Arnesano, F., Banci, L., Bertini, I., Ciofi-Baffoni, S., Woodyear, T.L., Johnson, C.M., Barker, P.D.(2000) Biochemistry 39: 1499-1514
- PubMed: 10684632
- DOI: https://doi.org/10.1021/bi991831o
- Primary Citation of Related Structures:
1QQ3 - PubMed Abstract:
An NMR characterization of the 98Arg --> Cys variant of iron (III)-containing cytochrome b562 from Escherichia coli has been performed and the solution structure obtained. This variant has a covalent bond between the heme and Cys 98, thus mimicking the heme binding in cytochrome c. The R98C cytochrome is shown to have a significantly increased stability, compared to that of wild type, toward thermal and chemical denaturation. In water at 20 degrees C it is 5.60 kJ mol-1 more stable than the WT protein, measured by equilibrium guanidine hydrochloride denaturation. The structure has been obtained through two-dimensional total correlation spectroscopy (TOCSY) and nuclear Overhauser effect spectroscopy (NOESY) experiments and through three-dimensional NOESY-15N heteronuclear multiple quantum coherence (HMQC). By these methods, 85% of protons and 100% of backbone nitrogens were assigned. 2145 meaningful nuclear Overhauser effects (NOEs) (20 NOEs per residue), 45 backbone 3J values, and 397 pseudocontact shifts were used to obtain a family of 35 members, which were then energy-minimized. The root-mean-square deviation (RMSD) with respect to the average structure is 0.50 +/- 0.07 for the backbone and 1.01 +/- 0.08 for the heavy atoms. The magnetic anisotropy resulting from analysis of the pseudocontact shifts indicates an anisotropy that is an intermediate between that of the wild-type, which is the smallest, and cytochrome c. The g values confirm a higher anisotropy of the variant with respect to the wild-type protein. The chirality of the heme 2 alpha carbon is the same as that in all naturally occurring cytochromes c. The overall secondary structure and tertiary structure are very similar to the wild type. The removal of Arg 98 causes a change in the pH-dependent properties. The pKa, proposed to be due to deprotonation of the coordinated histidine, is 1.5 units higher than in the wild type, consistent with the lack of the positive charge of Arg 98 close to the ionizable group. This is further support for the coordinated histidine being the titratable group with an alkaline pKa in the wild-type protein. The pattern of the shifts of the heme methyl groups is different than in the wild-type protein, presumably due to alteration of the electronic structure by the presence of the covalent bond between the protein and the heme. The difference in stability between the variant and wild-type protein is discussed in terms of the structural information.
Organizational Affiliation:
Centre for Protein Engineering, MRC Centre, Hills Road, Cambridge CB2 2QH, United Kingdom.