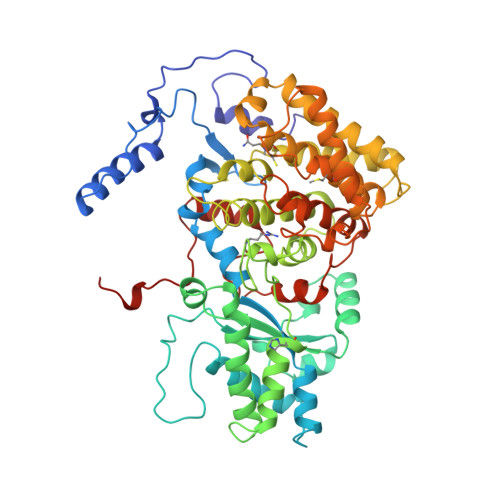
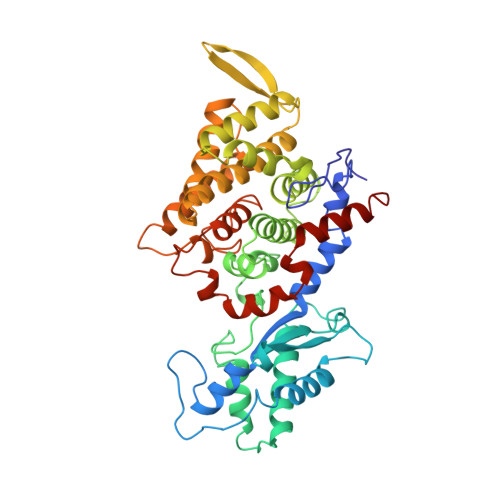
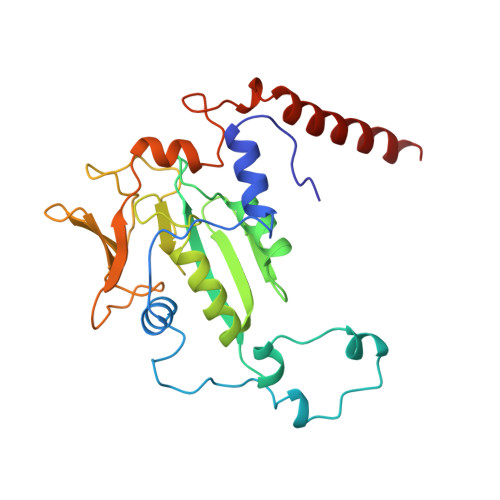
Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation.
Ermler, U., Grabarse, W., Shima, S., Goubeaud, M., Thauer, R.K.(1997) Science 278: 1457-1462
- PubMed: 9367957
- DOI: https://doi.org/10.1126/science.278.5342.1457
- Primary Citation of Related Structures:
1MRO - PubMed Abstract:
Methyl-coenzyme M reductase (MCR), the enzyme responsible for the microbial formation of methane, is a 300-kilodalton protein organized as a hexamer in an alpha2beta2gamma2 arrangement. The crystal structure of the enzyme from Methanobacterium thermoautotrophicum, determined at 1.45 angstrom resolution for the inactive enzyme state MCRox1-silent, reveals that two molecules of the nickel porphinoid coenzyme F430 are embedded between the subunits alpha, alpha', beta, and gamma and alpha', alpha, beta', and gamma', forming two identical active sites. Each site is accessible for the substrate methyl-coenzyme M through a narrow channel locked after binding of the second substrate coenzyme B. Together with a second structurally characterized enzyme state (MCRsilent) containing the heterodisulfide of coenzymes M and B, a reaction mechanism is proposed that uses a radical intermediate and a nickel organic compound.
Organizational Affiliation:
Max-Planck-Institut für Biophysik, Heinrich-Hoffmann-Strabetae 7, 60528 Frankfurt, Germany.