Structure of ribosomal protein TL5 complexed with RNA provides new insights into the CTC family of stress proteins.
Fedorov, R., Meshcheryakov, V., Gongadze, G., Fomenkova, N., Nevskaya, N., Selmer, M., Laurberg, M., Kristensen, O., Al-Karadaghi, S., Liljas, A., Garber, M., Nikonov, S.(2001) Acta Crystallogr D Biol Crystallogr 57: 968-976
- PubMed: 11418764
- DOI: https://doi.org/10.1107/s0907444901006291
- Primary Citation of Related Structures:
1FEU - PubMed Abstract:
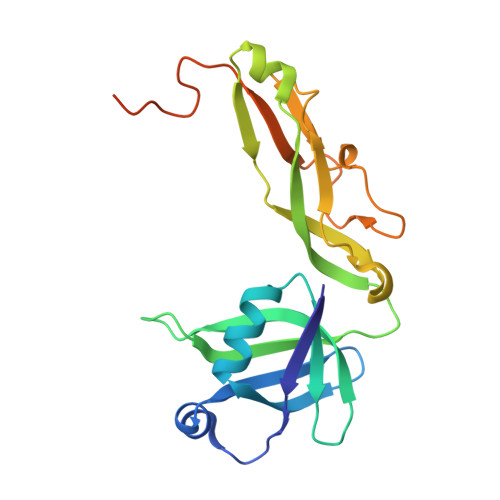
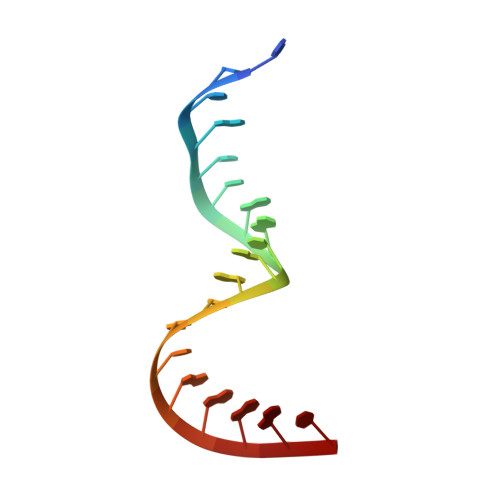
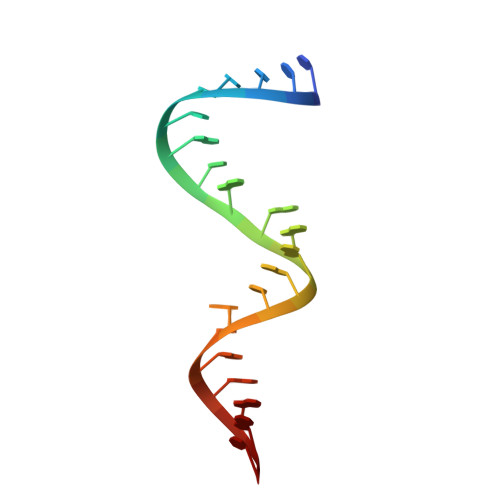
The crystal structure of Thermus thermophilus ribosomal protein TL5 in complex with a fragment of Escherichia coli 5S rRNA has been determined at 2.3 A resolution. The protein consists of two domains. The structure of the N-terminal domain is close to the structure of E. coli ribosomal protein L25, but the C-terminal domain represents a new fold composed of seven beta-strands connected by long loops. TL5 binds to the RNA through its N-terminal domain, whereas the C-terminal domain is not included in this interaction. Cd(2+) ions, the presence of which improved the crystal quality significantly, bind only to the protein component of the complex and stabilize the protein molecule itself and the interactions between the two molecules in the asymmetric unit of the crystal. The TL5 sequence reveals homology to the so-called general stress protein CTC. The hydrophobic cores which stabilize both TL5 domains are highly conserved in CTC proteins. Thus, all CTC proteins may fold with a topology close to that of TL5.
Organizational Affiliation:
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia.