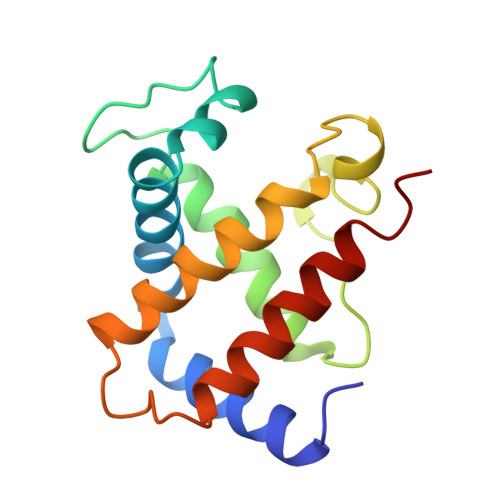
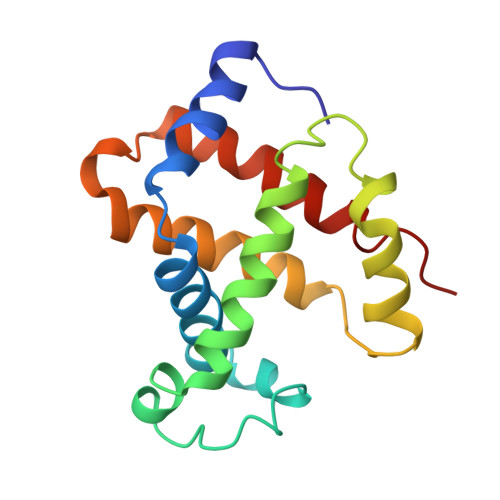
Crystal structure of Lysbeta(1)82-Lysbeta(2)82 crosslinked hemoglobin: a possible allosteric intermediate
Fernandez, E.J., Abad-Zapatero, C., Olsen, K.W.(2000) J Mol Biol 296: 1245-1256
- PubMed: 10698631
- DOI: https://doi.org/10.1006/jmbi.2000.3525
- Primary Citation of Related Structures:
1BIJ - PubMed Abstract:
The crystal structure of human hemoglobin crosslinked between the Lysbeta82 residues has been determined at 2.30 A resolution. The crosslinking reaction was performed under oxy conditions using bis(3, 5-dibromosalicyl) fumarate; the modified hemoglobin has increased oxygen affinity and lacks cooperativity. Since the crystallization occurred under deoxy conditions, the resulting structure displays conformational characteristics of both the (oxy) R and the (deoxy) T-states. beta82XLHbA does not fully reach its T-state conformation due to the presence of the crosslink. The R-state-like characteristics of deoxy beta82XLHbA include the position of the distal Hisbeta63 (E7) residue, indicating a possible reason for the high oxygen affinity of this derivative. Other areas of the molecule, particularly those thought to be important in the allosteric transition, such as Tyrbeta145 (HC2) and the switch region involving Proalpha(1)44 (CD2), Thralpha(1)41 (C6) and Hisbeta(2)97 (FG4), are in intermediate positions between the R and T-states. Thus, the structure may represent a stabilized intermediate in the allosteric transition of hemoglobin.
Organizational Affiliation:
Department of Chemistry, Loyola University Chicago, Chicago, IL 60626, USA.