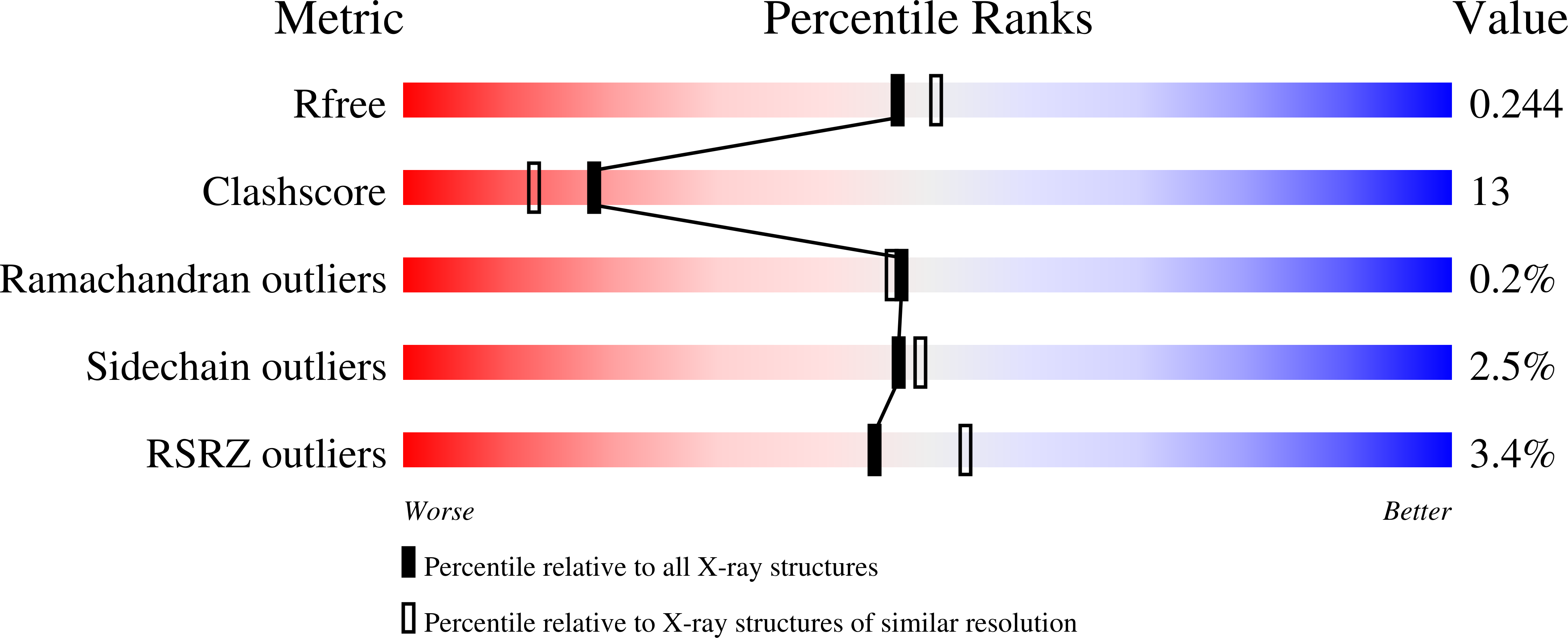
X-ray diffraction structure of a cell-wall invertase from Arabidopsis thaliana.
Verhaest, M., Lammens, W., Le Roy, K., De Coninck, B., De Ranter, C.J., Van Laere, A., Van den Ende, W., Rabijns, A.(2006) Acta Crystallogr D Biol Crystallogr 62: 1555-1563
- PubMed: 17139091
- DOI: https://doi.org/10.1107/S0907444906044489
- Primary Citation of Related Structures:
2AC1 - PubMed Abstract:
Cell-wall invertases play crucial roles during plant development. They hydrolyse sucrose into its fructose and glucose subunits by cleavage of the alpha1-beta2 glycosidic bond. Here, the structure of the Arabidopsis thaliana cell-wall invertase 1 (AtcwINV1; gene accession code At3g13790) is described at a resolution of 2.15 A. The structure comprises an N-terminal fivefold beta-propeller domain followed by a C-terminal domain formed by two beta-sheets. The active site is positioned in the fivefold beta-propeller domain, containing the nucleophile Asp23 and the acid/base catalyst Glu203 of the double-displacement enzymatic reaction. The function of the C-terminal domain remains unknown. Unlike in other GH 32 family enzyme structures known to date, in AtcwINV1 the cleft formed between both domains is blocked by Asn299-linked carbohydrates. A preliminary site-directed mutagenesis experiment (Asn299Asp) removed the glycosyl chain but did not alter the activity profile of the enzyme.
Organizational Affiliation:
Laboratorium voor Biokristallografie, Faculteit Farmaceutische Wetenschappen, K. U. Leuven, Herestraat 49, O&N II, Bus 822, B-3000 Leuven, Belgium.